Vesiculoviral matrix (M) protein occupies nucleic acid binding site at nucleoporin pair (Rae1 • Nup98) - PubMed (original) (raw)
Vesiculoviral matrix (M) protein occupies nucleic acid binding site at nucleoporin pair (Rae1 • Nup98)
Beili Quan et al. Proc Natl Acad Sci U S A. 2014.
Abstract
mRNA export factor 1 (Rae1) and nucleoporin 98 (Nup98) are host cell targets for the matrix (M) protein of vesicular stomatitis virus (VSV). How Rae1 functions in mRNA export and how M protein targets both Rae1 and Nup98 are not understood at the molecular level. To obtain structural insights, we assembled a 1:1:1 complex of M•Rae1•Nup98 and established a crystal structure at 3.15-Å resolution. We found that the M protein contacts the Rae1•Nup98 heterodimer principally by two protrusions projecting from the globular domain of M like a finger and thumb. Both projections clamp to the side of the β-propeller of Rae1, with the finger also contacting Nup98. The most prominent feature of the finger is highly conserved Methionine 51 (Met51) with upstream and downstream acidic residues. The complementary surface on Rae1 displays a deep hydrophobic pocket, into which Met51 fastens like a bolt, and a groove of basic residues on either side, which bond to the acidic residues of the finger. Notably, the M protein competed for in vitro binding of various oligonucleotides to Rae1•Nup98. We localized this competing activity of M to its finger using a synthetic peptide. Collectively, our data suggest that Rae1 serves as a binding protein for the phosphate backbone of any nucleic acid and that the finger of M mimics this ligand. In the context of mRNA export, we propose that a given mRNA segment, after having been deproteinated by helicase, is transiently reproteinated by Nup98-tethered Rae1. We suggest that such repetitive cycles provide cytoplasmic stopover sites required for ratcheting mRNA across the nuclear pore.
Keywords: M protein mimicry; mRNP export; phosphate backbone binding.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Fig. 1.
Structure of a 1:1:1 complex of M•Rae1•Nup98. (A) Domains of each full-length polypeptide are annotated (
Figs. S1
–
S3
). Recombinant protein fragments that were used for assembling the M•Rae1•Nup98 trimeric complex for crystallization (in the text) are indicated by bars and color-coded: blue, Rae1; magenta, Nup98; yellow, M protein. Moreover, the three surface features of M protein interacting with Rae1•Nup98 (see below) were designated and color-coded: brown, web (residues 142–144); green, thumb (residues 213–223); orange, finger (residues 49–61). Nup98 features numerous FG repeats, a predicted unstructured region, and binding sites for Rae1 (also referred to as GLEBS for Gle2 binding sequence) and Nup88 (denoted by NPC anchor) (Fig. 5). (B) Schematic topology diagram: location of secondary structural elements of the globular portion of M protein with respect to finger, web, and thumb. (C) Schematic contour diagram of the trimeric complex colored as in A and viewed as in D, Upper. (D) Ribbon presentation of the trimeric complex, colored as in A; Upper and Lower represent top and side views, respectively, with regard to the β-propeller domain of Rae1 and include annotations for secondary and tertiary structures of the three moieties of the trimeric complex (
Figs. S1
–
S3
). Note that the finger of M protein contacts both Rae1 and Nup98 moieties, whereas the web and thumb contact only Rae1.
Fig. 2.
Interface between M protein and Rae1•Nup98. (A) All residues involved at the interface between M and Rae1•Nup98 are shown. Black dotted lines indicate polar interactions, including salt bridges and hydrogen bonds. Gray lines indicate van der Waals (VDW) interactions. (B and C) Ribbon representation is colored as in Fig. 1. B, Inset and C, Inset illustrate the positions of (B) index finger and (C) thumb and web of the M protein, respectively and are expanded B, Lower and C, Lower. Polar interaction network at the interfaces is indicated by black dashes.
Fig. 3.
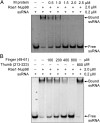
VSV M protein displaces ssRNA from Rae1FL•Nup98. (A) After preincubation of Alexa-488–labeled 10-mer poly(U) ssRNA and Rae1FL•Nup98157–213, the mixture was further incubated with increasing amounts of *M44–229 and analyzed by an EMSA. The final concentrations of the mixture are indicated. Presence of RNA was detected by fluorescence of Alexa-488. (B) Increasing amounts of the 13-mer finger peptide (residues 49–61) as indicated were added to the preformed ssRNA•Rae1FL•Nup98157–213 complex and analyzed as in A. The 11-mer thumb peptide (residues 213–223) was tested at 800 μM.
Fig. 4.
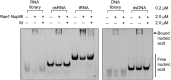
VSV M protein displaces different types of nucleic acids from the Rae1•Nup98 complex. After preincubation with nucleic acid and the Rae1FL•Nup98157–213 complex, the mixture was further incubated in the absence or presence of *M44–229. All final concentrations are indicated on the right. Representative nucleic acids include 21-mer RNA and DNA libraries containing random sequences, 21-mer dsRNA and dsDNA of nonrandom sequence (28), and yeast tRNAs (Roche). After EMSA, nucleic acid binding was visualized by SYBR-GOLD nucleic acid staining. Note that *M44–229 displaced each of the tested nucleic acids from the Rae1FL•Nup98157–213 complex.
Fig. 5.
Neighborhood of Rae1•Nup98 on the cytoplasmic side of the nuclear pore. (A) An overall schematic model for NPC architecture. It shows a series of concentric cylinders (anchored to the pore membrane domain of the nuclear envelope) and the attached cytoplasmic filaments and nuclear basket. A recently proposed structural model for the central transport channel with a midplane ring (shown in the open state) and attached structures on either side (16) are indicated. Modified from ref. . (B–E) Close-up views of components of one cytoplasmic filament (shaded in pink in A): the crystal structures are composites representing interactomes of either yeast or vertebrate (in parentheses) nucleoporins, color-coded, and shown in (B) space-filling or (C–E) ribbon representation. (C) yGle1•Dbp5•Nup159 [Protein Data Bank (PDB) ID code 3RRM] (24). (D) Human Rae1•Nup98 (PDB ID code 4OWR). (E) yNup82•Nup159•Nup116 (PDB ID code 3PBP) (21). At the cytoplasmic filament base, yNup82 (Nup88) serves as a node. (B and E) Its N-terminal domain interactome encompasses the C-terminal tail of yNup159 and the C-terminal domain (CTD) of yNup116 (Nup98). (B) The yNup82 (Nup88)-CTD interactome with nucleoporin(s) of the cylindrical core still awaits analyses at the molecular level. The farther course of yNup159 (Nup214) and yNup116 (Nup98) with (B–E) upstream interactomes and various unstructured regions with and without FG repeats is annotated in B (in the text); the length of the color-coded lines is roughly proportional to the predicted length of unstructured regions. Note that five homodimers of yeast dynein light chain (yDyn2) link two yNup159 molecules and thereby, dimerize two yNup82 modules. Because interactomes of vertebrate Nup358 and hCG1 (yNup42) with other nucleoporins remain uncharacterized at the molecular level, they have been omitted in this neighborhood model of Rae1•Nup98.
Fig. 6.
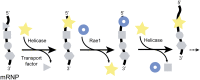
Rae1•Nup98 functions as a phosphate backbone-binding protein in a ratchet model for mRNA export across the central transport channel of the nuclear pore. At cytoplasmic filaments of the nuclear pore, mRNA-bound proteins (gray symbols) are sequentially removed, one by one, in the 5′ to 3′ direction (29). The removal of each protein requires one ATPase cycle of helicase (yellow star) (24, 30, 31). The protein-vacated stretch of the mRNA phosphate backbone is immediately occupied by the nearby Rae1 (blue ring) (in the text and Fig. 5). The number of these repeat cycles is predicted to be proportional to the length of mRNA.
Similar articles
- SARS-CoV-2 ORF6 Disrupts Bidirectional Nucleocytoplasmic Transport through Interactions with Rae1 and Nup98.
Addetia A, Lieberman NAP, Phung Q, Hsiang TY, Xie H, Roychoudhury P, Shrestha L, Loprieno MA, Huang ML, Gale M Jr, Jerome KR, Greninger AL. Addetia A, et al. mBio. 2021 Apr 13;12(2):e00065-21. doi: 10.1128/mBio.00065-21. mBio. 2021. PMID: 33849972 Free PMC article. - Structural and functional analysis of the interaction between the nucleoporin Nup98 and the mRNA export factor Rae1.
Ren Y, Seo HS, Blobel G, Hoelz A. Ren Y, et al. Proc Natl Acad Sci U S A. 2010 Jun 8;107(23):10406-11. doi: 10.1073/pnas.1005389107. Epub 2010 May 24. Proc Natl Acad Sci U S A. 2010. PMID: 20498086 Free PMC article. - Complexes of vesicular stomatitis virus matrix protein with host Rae1 and Nup98 involved in inhibition of host transcription.
Rajani KR, Pettit Kneller EL, McKenzie MO, Horita DA, Chou JW, Lyles DS. Rajani KR, et al. PLoS Pathog. 2012 Sep;8(9):e1002929. doi: 10.1371/journal.ppat.1002929. Epub 2012 Sep 27. PLoS Pathog. 2012. PMID: 23028327 Free PMC article. - Nucleoporin Nup98: a gatekeeper in the eukaryotic kingdoms.
Iwamoto M, Asakawa H, Hiraoka Y, Haraguchi T. Iwamoto M, et al. Genes Cells. 2010 Jun;15(7):661-9. doi: 10.1111/j.1365-2443.2010.01415.x. Epub 2010 Jun 7. Genes Cells. 2010. PMID: 20545767 Review. - Nucleoporins and nucleocytoplasmic transport in hematologic malignancies.
Takeda A, Yaseen NR. Takeda A, et al. Semin Cancer Biol. 2014 Aug;27:3-10. doi: 10.1016/j.semcancer.2014.02.009. Epub 2014 Mar 18. Semin Cancer Biol. 2014. PMID: 24657637 Review.
Cited by
- The Structure of the Nuclear Pore Complex (An Update).
Lin DH, Hoelz A. Lin DH, et al. Annu Rev Biochem. 2019 Jun 20;88:725-783. doi: 10.1146/annurev-biochem-062917-011901. Epub 2019 Mar 18. Annu Rev Biochem. 2019. PMID: 30883195 Free PMC article. Review. - SARS-CoV-2 Orf6 is positioned in the nuclear pore complex by Rae1 to inhibit nucleocytoplasmic transport.
Makio T, Zhang K, Love N, Mast FD, Liu X, Elaish M, Hobman T, Aitchison JD, Fontoura BMA, Wozniak RW. Makio T, et al. Mol Biol Cell. 2024 May 1;35(5):ar62. doi: 10.1091/mbc.E23-10-0386. Epub 2024 Mar 20. Mol Biol Cell. 2024. PMID: 38507240 Free PMC article. - SARS-CoV-2 ORF6 Disrupts Bidirectional Nucleocytoplasmic Transport through Interactions with Rae1 and Nup98.
Addetia A, Lieberman NAP, Phung Q, Hsiang TY, Xie H, Roychoudhury P, Shrestha L, Loprieno MA, Huang ML, Gale M Jr, Jerome KR, Greninger AL. Addetia A, et al. mBio. 2021 Apr 13;12(2):e00065-21. doi: 10.1128/mBio.00065-21. mBio. 2021. PMID: 33849972 Free PMC article. - Proteomic elucidation of the targets and primary functions of the picornavirus 2A protease.
Serganov AA, Udi Y, Stein ME, Patel V, Fridy PC, Rice CM, Saeed M, Jacobs EY, Chait BT, Rout MP. Serganov AA, et al. J Biol Chem. 2022 Jun;298(6):101882. doi: 10.1016/j.jbc.2022.101882. Epub 2022 Mar 31. J Biol Chem. 2022. PMID: 35367208 Free PMC article. - Structural basis for Sarbecovirus ORF6 mediated blockage of nucleocytoplasmic transport.
Gao X, Tian H, Zhu K, Li Q, Hao W, Wang L, Qin B, Deng H, Cui S. Gao X, et al. Nat Commun. 2022 Aug 15;13(1):4782. doi: 10.1038/s41467-022-32489-5. Nat Commun. 2022. PMID: 35970938 Free PMC article.
References
- Faria PA, et al. VSV disrupts the Rae1/mrnp41 mRNA nuclear export pathway. Mol Cell. 2005;17(1):93–102. - PubMed
- von Kobbe C, et al. Vesicular stomatitis virus matrix protein inhibits host cell gene expression by targeting the nucleoporin Nup98. Mol Cell. 2000;6(5):1243–1252. - PubMed
- Hoelz A, Debler EW, Blobel G. The structure of the nuclear pore complex. Annu Rev Biochem. 2011;80:613–643. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources