Probing nuclear pore complex architecture with proximity-dependent biotinylation - PubMed (original) (raw)
Probing nuclear pore complex architecture with proximity-dependent biotinylation
Dae In Kim et al. Proc Natl Acad Sci U S A. 2014.
Abstract
Proximity-dependent biotin identification (BioID) is a method for identifying protein associations that occur in vivo. By fusing a promiscuous biotin ligase to a protein of interest expressed in living cells, BioID permits the labeling of proximate proteins during a defined labeling period. In this study we used BioID to study the human nuclear pore complex (NPC), one of the largest macromolecular assemblies in eukaryotes. Anchored within the nuclear envelope, NPCs mediate the nucleocytoplasmic trafficking of numerous cellular components. We applied BioID to constituents of the Nup107-160 complex and the Nup93 complex, two conserved NPC subcomplexes. A strikingly different set of NPC constituents was detected depending on the position of these BioID-fusion proteins within the NPC. By applying BioID to several constituents located throughout the extremely stable Nup107-160 subcomplex, we refined our understanding of this highly conserved subcomplex, in part by demonstrating a direct interaction of Nup43 with Nup85. Furthermore, by using the extremely stable Nup107-160 structure as a molecular ruler, we defined the practical labeling radius of BioID. These studies further our understanding of human NPC organization and demonstrate that BioID is a valuable tool for exploring the constituency and organization of large protein assemblies in living cells.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
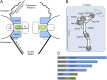
Fig. 1.
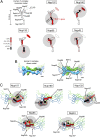
Organization of the mammalian NPC, Y-complex, and BirA*-fusion proteins. (A) Positioning of the Y-complex (blue) and Nup53–93 complex (green) within a simplified model of NPC organization. A full description of the members of each pore subcomplex is shown in Table 1, leftmost column. TM Nups, transmembrane Nups. (B) Model of the human Y-complex. Its many β-propeller domains are schematized by circles or bulges; alpha-solenoid folds are depicted by rectangles. Nup43 is drawn with a dashed line because its localization was unknown at the time these studies were performed. The dotted line and oval indicates possible residence of ELYS near Nup160 and Nup37, extrapolated from studies in yeast (63, 64). Gray disks represent the predicted localization of the BirA*-ligase based on available structural data. (C) Linear model for the NPC proteins fused to BirA* for the BioID studies.
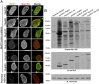
Fig. 2.
Characterization of cells stably expressing NPC BirA*-fusion proteins. (A) Immunofluorescence analyses of HEK293 cells stably expressing Y-complex members or Nup53 fused to BirA*. The biotin signal generated by the BirA*-fusion proteins is detected with fluorescently labeled streptavidin (green). and NPCs are detected with anti-Nup153 (red). (Scale bar: 7 μm.) (B) Following SDS/PAGE of cell lysates, biotinylated proteins were detected with streptavidin-HRP. Asterisks indicate the location of the BirA*-fusion protein (detected with anti-BirA). Tubulin was used as loading control.
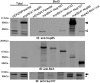
Fig. 3.
Nup85 is a poor substrate for BioID. IB analysis detects low levels of endogenous Nup85 (open arrowhead) in the Nup160, Nup107, and Nup43 BioID pull-down samples and significant levels of the exogenous mycBirA*-Nup85 (arrowheads) in the Nup85 BioID pull-down. (Top) For clarity, total lysates are shown at a lower exposure than the BioID samples. (Middle) For comparison, we detect similar levels of the BirA*-fusion proteins with anti-BirA in these same samples. (Bottom) Reprobing the same membrane with anti-Nup107 reveals levels of endogenous Nup107 (arrow) that correlate with the MS results, thus corroborating those results. Exogenous mycBirA*-Nup85 remains detected below the endogenous Nup107 (asterisk).
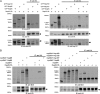
Fig. 4.
Nup43 interacts with Nup85. (A) Anti-HA co-IP from lysates of HEK293 cells cotransfected with Nup43-HA (Middle, arrowhead) and GFP–Y-Nups (Top) indicates that GFP-Nup85 is pulled down most efficiently by Nup43-HA. (Bottom) Reprobing the samples with anti-Nup107 reveals low levels of endogenous Nup107 (arrow) in all the Nup43-HA pull-down samples. (B) Co-IPs from in vitro transcription/translation reactions in reticulocyte lysates using Nup43-HA (Lower, arrowhead) alone or with mycBirA*-tagged Y-Nups (Upper). Only mycBirA*-Nup85 is detected in the Nup43-HA pull-down fraction.
Fig. 5.
Biotinylation of Y-Nups in the context of the whole NPC defines a practical labeling radius. (A) For each BioID-fusion, a model of a single Y-complex subunit is used to depict the relative abundance of Y-Nups detected following BioID pull-down. The red circles depict the approximate position of the BirA* ligase. Gray disks (10-nm radius) provide an approximation of the labeling radius of BioID. (B) Structural model from Bui et al. (21) in which offset Y-complex dimers are arranged in a head-to-tail fashion within the NPC (Left). The approximate positions of Y-Nups are labeled and schematized (Right) on this map. (Modified from ref. .) (C) BioID data were applied to the dimer model of Y-complex. The color code in A is used to depict the relative abundance of biotinylated Y-Nups for BioID-fusion proteins. The gray disks (dark: 5-nm radius; light: 10-nm radius) provide an approximation of the labeling radius of BioID.
Fig. 6.
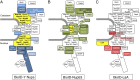
Biotinylation of NPC constituents generally correlates with the location of the fusion protein. The candidate Nups identified in this BioID studies are positioned within a simplified model of NPC organization that integrates data from the literature and extrapolations based on previous studies in budding yeast. The baits (bold text) used are shaded yellow. Intensity of (A) blue- (BioID-Y-Nups), (B) green- (BioID-Nup53), or (C) red- (BioID-LaA) shaded candidates correlates with the level of detection of candidates predominantly detected by the different types of BioID-fusion proteins (Table 1). In A, biotinylated Y-Nups are not shaded blue for clarity (Fig. 5). The asterisks next to Nup62 in A and next to Nup133, Nup96, and Nup98 in C represent candidates with multiple locations within the NPC and are unlikely to be biotinylated at that specific place.
Similar articles
- Nuclear pore complexes undergo Nup221 exchange during blood-stage asexual replication of Plasmodium parasites.
Blauwkamp J, Ambekar SV, Hussain T, Mair GR, Beck JR, Absalon S. Blauwkamp J, et al. mSphere. 2024 Dec 19;9(12):e0075024. doi: 10.1128/msphere.00750-24. Epub 2024 Nov 11. mSphere. 2024. PMID: 39526784 Free PMC article. - Nuclear basket proteins regulate the distribution and mobility of nuclear pore complexes in budding yeast.
Zsok J, Simon F, Bayrak G, Isaki L, Kerff N, Kicheva Y, Wolstenholme A, Weiss LE, Dultz E. Zsok J, et al. Mol Biol Cell. 2024 Nov 1;35(11):ar143. doi: 10.1091/mbc.E24-08-0371. Epub 2024 Sep 25. Mol Biol Cell. 2024. PMID: 39320946 Free PMC article. - Defining the optimum strategy for identifying adults and children with coeliac disease: systematic review and economic modelling.
Elwenspoek MM, Thom H, Sheppard AL, Keeney E, O'Donnell R, Jackson J, Roadevin C, Dawson S, Lane D, Stubbs J, Everitt H, Watson JC, Hay AD, Gillett P, Robins G, Jones HE, Mallett S, Whiting PF. Elwenspoek MM, et al. Health Technol Assess. 2022 Oct;26(44):1-310. doi: 10.3310/ZUCE8371. Health Technol Assess. 2022. PMID: 36321689 Free PMC article. - Depressing time: Waiting, melancholia, and the psychoanalytic practice of care.
Salisbury L, Baraitser L. Salisbury L, et al. In: Kirtsoglou E, Simpson B, editors. The Time of Anthropology: Studies of Contemporary Chronopolitics. Abingdon: Routledge; 2020. Chapter 5. In: Kirtsoglou E, Simpson B, editors. The Time of Anthropology: Studies of Contemporary Chronopolitics. Abingdon: Routledge; 2020. Chapter 5. PMID: 36137063 Free Books & Documents. Review. - The effectiveness of abstinence-based and harm reduction-based interventions in reducing problematic substance use in adults who are experiencing homelessness in high income countries: A systematic review and meta-analysis: A systematic review.
O'Leary C, Ralphs R, Stevenson J, Smith A, Harrison J, Kiss Z, Armitage H. O'Leary C, et al. Campbell Syst Rev. 2024 Apr 21;20(2):e1396. doi: 10.1002/cl2.1396. eCollection 2024 Jun. Campbell Syst Rev. 2024. PMID: 38645303 Free PMC article. Review.
Cited by
- CLK1/CLK2-driven signalling at the Leishmania kinetochore is captured by spatially referenced proximity phosphoproteomics.
Geoghegan V, Carnielli JBT, Jones NG, Saldivia M, Antoniou S, Hughes C, Neish R, Dowle A, Mottram JC. Geoghegan V, et al. Commun Biol. 2022 Nov 28;5(1):1305. doi: 10.1038/s42003-022-04280-1. Commun Biol. 2022. PMID: 36437406 Free PMC article. - Determination of local chromatin composition by CasID.
Schmidtmann E, Anton T, Rombaut P, Herzog F, Leonhardt H. Schmidtmann E, et al. Nucleus. 2016 Sep 2;7(5):476-484. doi: 10.1080/19491034.2016.1239000. Epub 2016 Sep 27. Nucleus. 2016. PMID: 27676121 Free PMC article. - Comparative analysis of retroviral Gag-host cell interactions: focus on the nuclear interactome.
Lambert GS, Rice BL, Kaddis Maldonado RJ, Chang J, Parent LJ. Lambert GS, et al. bioRxiv [Preprint]. 2024 Mar 6:2024.01.18.575255. doi: 10.1101/2024.01.18.575255. bioRxiv. 2024. PMID: 38293010 Free PMC article. Updated. Preprint. - Identifying and Validating MYC:Protein Interactors in Pursuit of Novel Anti-MYC Therapies.
Resetca D, MacDonald AS, Kenney TMG, Wei Y, Arrowsmith CH, Raught B, Penn LZ. Resetca D, et al. Methods Mol Biol. 2021;2318:45-67. doi: 10.1007/978-1-0716-1476-1_4. Methods Mol Biol. 2021. PMID: 34019286 - Force dependent biotinylation of myosin IIA by α-catenin tagged with a promiscuous biotin ligase.
Ueda S, Blee AM, Macway KG, Renner DJ, Yamada S. Ueda S, et al. PLoS One. 2015 Mar 25;10(3):e0122886. doi: 10.1371/journal.pone.0122886. eCollection 2015. PLoS One. 2015. PMID: 25806963 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
- P20GM103620/GM/NIGMS NIH HHS/United States
- R01EB014869/EB/NIBIB NIH HHS/United States
- R01 GM102486/GM/NIGMS NIH HHS/United States
- R01 EB014869/EB/NIBIB NIH HHS/United States
- P20 GM103620/GM/NIGMS NIH HHS/United States
- R01 GM102203/GM/NIGMS NIH HHS/United States
- R01GM102486/GM/NIGMS NIH HHS/United States
- P20GM103548/GM/NIGMS NIH HHS/United States
- R01GM102203/GM/NIGMS NIH HHS/United States
- P20 GM103548/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous