Regulation of cell proliferation and migration by p62 through stabilization of Twist1 - PubMed (original) (raw)
Clinical Trial
. 2014 Jun 24;111(25):9241-6.
doi: 10.1073/pnas.1322913111. Epub 2014 Jun 9.
Affiliations
- PMID: 24927592
- PMCID: PMC4078859
- DOI: 10.1073/pnas.1322913111
Clinical Trial
Regulation of cell proliferation and migration by p62 through stabilization of Twist1
Lei Qiang et al. Proc Natl Acad Sci U S A. 2014.
Retraction in
- Retraction for Qiang et al., Regulation of cell proliferation and migration by p62 through stabilization of Twist1.
[No authors listed] [No authors listed] Proc Natl Acad Sci U S A. 2023 Sep 5;120(36):e2313213120. doi: 10.1073/pnas.2313213120. Epub 2023 Aug 28. Proc Natl Acad Sci U S A. 2023. PMID: 37639612 Free PMC article. No abstract available.
Abstract
The selective autophagy substrate p62 serves as a molecular link between autophagy and cancer. Suppression of autophagy causes p62 accumulation and thereby contributes to tumorigenesis. Here we demonstrate that autophagy deficiency promotes cell proliferation and migration through p62-dependent stabilization of the oncogenic transcription factor Twist1. p62 binds to Twist1 and inhibits degradation of Twist1. In mice, p62 up-regulation promotes tumor cell growth and metastasis in a Twist1-dependent manner. Our findings demonstrate that Twist1 is a key downstream effector of p62 in regulation of cell proliferation and migration and suggest that targeting p62-mediated Twist1 stabilization is a promising therapeutic strategy for prevention and treatment of cancer.
Keywords: SQSTM1; melanoma; proteasome; skin cancer; ubiquitination.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Fig. 1.
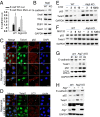
Autophagy deficiency decreases E-cadherin expression and promotes cell migration, invasion, and proliferation. (A) Immunoblot analysis of E-cadherin, p62, LC3-I/II, and GAPDH in WT and Atg3, Atg5, Atg9, and Atg12 KO MEF cells. (B) Transwell assay of the invasion ability of WT and Atg3, Atg5, Atg9, and Atg12 KO MEF cells. Migrated cells on the underside of Transwell filters were stained and observed under microscopy. (C) Radius migration assay of the migration ability of WT and Atg3, Atg5, Atg9, and Atg12 KO MEF cells. (D) CellTiter 96 AQueous Non-Radioactive Cell Proliferation Assay (MTS) of WT and Atg3, Atg5, Atg9, and Atg12 KO MEF cells. The results were obtained from three independent experiments [mean ± SD (error bars), n = 3; *P < 0.05; **P < 0.01, compared with the WT group]. (E) Representative histological and immunohistochemical analysis of p62 and E-cadherin protein levels (brown) in normal skin (n = 14), well-differentiated squamous cell carcinoma (SCC WD) (n = 25), and poorly differentiated SCC (SCC PD) (n = 10). Black squares indicate the region shown in higher magnification. (Scale bar, 200 μm and 50 μm for Left and Right, respectively.)
Fig. 2.
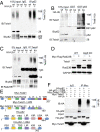
Autophagy deficiency delays Twist1 degradation via both autophagosome and proteasome pathways. (A) Luciferase reporter assay of the E-cadherin promoter with an intact (E-cad WT-Luc) or mutated (E-cad E-box Mut-Luc) E-box site in WT and Atg5 KO MEF cells. (B) Immunoblot analysis of N-cadherin, Snail, Slug, ZEB1, and Twist1 in WT and Atg5 KO MEF cells. (C and D) Immunofluorescence assay of the colocalization of p62 (C) or LC3 (D) with Twist1 in WT and Atg5 KO MEF cells following treatment with vehicle or rapamycin (500 nM) for 6 h. (Scale bar, 10 μm.) The blue is a DAPI nuclear counterstain. (E and F) Immunoblot analysis of p53, Twist1, and GAPDH in WT and Atg5 KO MEF cells treated with CHX (100 μg/mL, E) or MG132 (10 μM, F) for the indicated times. (G) Immunoblot analysis of p62, Twist1, and GAPDH in the skin from WT and Atg7 cKO mice. (H) Immunoblot analysis of E-cadherin, p62, Twist1, LC3-I/II, Atg7, and GAPDH in WT and Atg7 KO cells reconstituted with Atg7. These results were obtained from three independent experiments.
Fig. 3.
Autophagy deficiency stabilizes Twist1 via p62 accumulation. (A) Immunoblot analysis of E-cadherin, p62, Twist1, and GAPDH in WT and Atg5 KO cells transfected with vector and siRNA targeting p62 and Twist1. (B) Immunoblot analysis of E-cadherin, p62, Twist1, and GAPDH in WT and Atg5 KO cells transfected with vector and shRNA targeting p62 and Twist1. (C) Immunoblot analysis of p62, Twist1, and GAPDH in WT and Atg5 KO cells transfected with vector and p62. (D) Immunoblot analysis of p62, Twist1, and GAPDH in WT, p62 KO cells, and p62 KO cells reconstituted with p62. (E) Immunoblot analysis of HA, Myc, and GAPDH in 293T cells transfected with Myc-Twist1 and HA-p62 for 48 h and then treated with CHX (100 μg/mL) for the indicated times. These results were obtained from three independent experiments.
Fig. 4.
p62 binds to Twist1 through its UBA domain. (A) Immunoblot analysis of Twist1 and p62 following immunoprecipitation using control species-matched IgG and anti-p62 antibody in WT and Atg5 KO MEF cells. (B) GST pulldown assay of the binding of GST-p62 with Twist1 in WT and Atg5 KO MEF cells. (C) Immunoblot analysis of Twist1, p62, and Rad23B following immunoprecipitation using control species-matched IgG, and anti-Twist1 antibody in WT and Atg5 KO MEF cells. (D) Immunoblot analysis of Twist1, Rad23B, and GAPDH in WT and Atg5 KO MEF cells transfected with vector control or Myc-Flag-Rad23B. (E) Schematic for p62 and Twist1 deletion constructs. (F) Immunoblot analysis of HA (HA-p62) and Myc (Myc-Twist1) in total cell lysates (input) or HA (HA-Ub, and HA-p62) following immunoprecipitation using control species-matched IgG and anti-Myc (Myc-Twist1) antibody in WT and Atg5 KO MEF cells. Immunoprecipitation assay of the binding of p62 or p62-ΔUBA with Twist1 or Twist1-ΔWR in 293T cells transfected with the combination of Myc-Twist1 and HA-p62, the combination of Myc-Twist1 and HA-p62-ΔUBA, and the combination of HA-p62 and Myc-Twist1-ΔWR for 48 h. The results were obtained from three independent experiments. Molecular weight in kilodaltons is marked in Fig. 4 A_–_C and F.
Fig. 5.
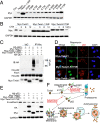
Twist1 lysine 175 is critical for Twist1 ubiquitination, degradation, and binding with p62. (A) Immunoblot analysis of Myc and GAPDH in 293T cells transfected with vector (Con), Myc-Twist1 (K→R) mutation constructs. (B) Immunoblot analysis of Myc and GAPDH in 293T cells transfected with Myc-Twist1, Myc-Twist1-ΔWR, or Myc-Twist1-K175R for 48 h and then treated with CHX (100 μg/mL) for the indicated times. (C) Immunoblot analysis of ubiquitinated Twist1 (HA) and HA-p62 using an anti-HA antibody following immunoprecipitation (IP) with an anti-Myc antibody, and Myc in 293T cells transfected with HA-Ub together with a combination of Myc-Twist1, Myc-Twist1-ΔWR, or Myc-Twist1-K175R with HA-p62. (D) Immunofluorescence assay of the colocalization of p62 and Myc in WT MEF cells transfected with Myc-Twist1, Myc-Twist1-ΔWR, or Myc-Twist1-K175R, following rapamycin (500 nM) treatment for 6 h. (Scale bar, 10 μm.) The blue is a DAPI nuclear counterstain. (E) Immunoblot analysis of E-cadherin, HA, and Myc in WT MEF cells transfected with Myc-Twist1, a combination of Myc-Twist1 and HA-p62, Myc-Twist1-ΔWR, Myc-Twist1-K175R, or a combination of Myc-Twist1-K175R and HA-p62. (F) Schematic for the regulation of Twist1 by p62 through autophagy and proteasome. These results were obtained from three independent experiments.
Fig. 6.
p62 promotes tumor cell growth and metastasis in mice through Twist1. (A) Immunoblot analysis of p62, Twist1, and GAPDH in A431 SCC cells transfected with vector, HA-p62, Twist1, or a combination of p62 and Twist1. (B) Luciferase reporter assay of the E-cadherin promoter with an intact (E-cad WT-Luc) or mutated (E-cad Mut-Luc) E-box site in A431 cells transfected with vector, p62, Twist1, or a combination of p62 and Twist1. (C) Wound healing assay of migration ability of A431 cells transfected with vector control (Con), Twist1, p62, or a combination of p62 and Twist1. (D) MTS assay of proliferation of A431 cells transfected with vector control (Con), p62, Twist1, or a combination of p62 and Twist1 as a function of days after transfection. (E) Average volume (mm3) of A431-Con, A431-p62, A431-Twist1, and A431-Twist1-p62 tumors at different weeks following s.c. injection. (F) Average number of lung tumor nodules per mouse in established A431-Twist1 and A431-Twist1-p62 lung metastasis tumors at 14 wk following injection. (G) Immunoblot analysis of HA, Twist1, and GAPDH in A375 melanoma cells transfected with vector or HA-p62. (H) Average volume (mm3) of A375-Con and A375-p62 tumors at different weeks following s.c. injection. (I) Immunoblot analysis of p62, Twist1, Atg7, and GAPDH in A375 melanoma cells transfected with vector or shRNA targeting Atg7 (shAtg7) or p62 (shp62). (J) Average volume (mm3) of A375-Con, A375-shp62, and A375-shAtg7 tumors at different weeks following s.c. injection. (K) Immunoblot analysis of E-cadherin, N-cadherin, Twist1, p62, and GAPDH in HaCaT human epithelial cells transfected with vector (shCon) or shp62, following treatment with EGF (10 ng/mL) together with TGF-β (10 ng/mL) (EGF/TGF-β) for 0, 6, 24, or 48 h. (L) Real-time RT-PCR analysis of Twist1 in HaCaT human epithelial cells transfected with shCon or shp62, following treatment with vehicle or EGF/TGF-β for 48 h. (M) Real-time RT-PCR analysis of p62 in HaCaT human epithelial cells following treatment with vehicle or EGF/TGF-β for 48 h. The results were obtained from three independent experiments [mean ± SD (error bars), n = 3; *P < 0.05; compared with WT Con cells (B); #P < 0.05; compared with the WT E-cadherin promoter (B); *P < 0.05; compared with the Con, A431-p62, and A431-Twist1 groups (E); and **P < 0.01; compared with the A431-Twist1 group (F)].
Similar articles
- Autophagy deficiency stabilizes TWIST1 to promote epithelial-mesenchymal transition.
Qiang L, He YY. Qiang L, et al. Autophagy. 2014 Oct 1;10(10):1864-5. doi: 10.4161/auto.32171. Epub 2014 Aug 13. Autophagy. 2014. PMID: 25126736 Free PMC article. - Translational downregulation of Twist1 expression by antiproliferative gene, B-cell translocation gene 2, in the triple negative breast cancer cells.
Devanand P, Sundaramoorthy S, Ryu MS, Jayabalan AK, Ohn T, Lim IK. Devanand P, et al. Cell Death Dis. 2019 May 28;10(6):410. doi: 10.1038/s41419-019-1640-z. Cell Death Dis. 2019. PMID: 31138781 Free PMC article. - Autophagy regulates tumor growth and metastasis.
Qiang L, Zhao B, Ming M, Wang N, He TC, Hwang S, Thorburn A, He YY. Qiang L, et al. bioRxiv [Preprint]. 2023 Nov 3:2023.10.31.564991. doi: 10.1101/2023.10.31.564991. bioRxiv. 2023. PMID: 37961427 Free PMC article. Preprint. - The interplay between microRNAs and Twist1 transcription factor: a systematic review.
Khanbabaei H, Teimoori A, Mohammadi M. Khanbabaei H, et al. Tumour Biol. 2016 Jun;37(6):7007-19. doi: 10.1007/s13277-016-4960-y. Epub 2016 Feb 15. Tumour Biol. 2016. PMID: 26880587 Review. - The role of TWIST1 in epithelial-mesenchymal transition and cancers.
Zhu QQ, Ma C, Wang Q, Song Y, Lv T. Zhu QQ, et al. Tumour Biol. 2016 Jan;37(1):185-97. doi: 10.1007/s13277-015-4450-7. Epub 2015 Nov 24. Tumour Biol. 2016. PMID: 26602382 Review.
Cited by
- Enhanced p62-NRF2 Feedback Loop due to Impaired Autophagic Flux Contributes to Arsenic-Induced Malignant Transformation of Human Keratinocytes.
Wu X, Sun R, Wang H, Yang B, Wang F, Xu H, Chen S, Zhao R, Pi J, Xu Y. Wu X, et al. Oxid Med Cell Longev. 2019 Oct 30;2019:1038932. doi: 10.1155/2019/1038932. eCollection 2019. Oxid Med Cell Longev. 2019. PMID: 31781319 Free PMC article. - TSSC3 promotes autophagy via inactivating the Src-mediated PI3K/Akt/mTOR pathway to suppress tumorigenesis and metastasis in osteosarcoma, and predicts a favorable prognosis.
Zhao GS, Gao ZR, Zhang Q, Tang XF, Lv YF, Zhang ZS, Zhang Y, Tan QL, Peng DB, Jiang DM, Guo QN. Zhao GS, et al. J Exp Clin Cancer Res. 2018 Aug 9;37(1):188. doi: 10.1186/s13046-018-0856-6. J Exp Clin Cancer Res. 2018. PMID: 30092789 Free PMC article. - Autophagy and cancer: Modulation of cell death pathways and cancer cell adaptations.
Towers CG, Wodetzki D, Thorburn A. Towers CG, et al. J Cell Biol. 2020 Jan 6;219(1):e201909033. doi: 10.1083/jcb.201909033. J Cell Biol. 2020. PMID: 31753861 Free PMC article. Review. - Loss of PGC-1α in RPE induces mesenchymal transition and promotes retinal degeneration.
Rosales MAB, Shu DY, Iacovelli J, Saint-Geniez M. Rosales MAB, et al. Life Sci Alliance. 2019 May 17;2(3):e201800212. doi: 10.26508/lsa.201800212. Print 2019 Jun. Life Sci Alliance. 2019. PMID: 31101737 Free PMC article. - Autophagic Degradation of NBR1 Restricts Metastatic Outgrowth during Mammary Tumor Progression.
Marsh T, Kenific CM, Suresh D, Gonzalez H, Shamir ER, Mei W, Tankka A, Leidal AM, Kalavacherla S, Woo K, Werb Z, Debnath J. Marsh T, et al. Dev Cell. 2020 Mar 9;52(5):591-604.e6. doi: 10.1016/j.devcel.2020.01.025. Epub 2020 Feb 20. Dev Cell. 2020. PMID: 32084360 Free PMC article.
References
- Klionsky DJ. Autophagy: From phenomenology to molecular understanding in less than a decade. Nat Rev Mol Cell Biol. 2007;8(11):931–937. - PubMed
- Choi AM, Ryter SW, Levine B. Autophagy in human health and disease. N Engl J Med. 2013;368(19):1845–1846. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- ES016936/ES/NIEHS NIH HHS/United States
- UL1 RR024999/RR/NCRR NIH HHS/United States
- P30 CA014599/CA/NCI NIH HHS/United States
- R01 ES016936/ES/NIEHS NIH HHS/United States
- UL1 TR000430/TR/NCATS NIH HHS/United States
- NIH UL1RR024999/RR/NCRR NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases