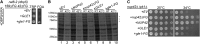Nucleoporin FG domains facilitate mRNP remodeling at the cytoplasmic face of the nuclear pore complex - PubMed (original) (raw)
Nucleoporin FG domains facilitate mRNP remodeling at the cytoplasmic face of the nuclear pore complex
Rebecca L Adams et al. Genetics. 2014 Aug.
Abstract
Directional export of messenger RNA (mRNA) protein particles (mRNPs) through nuclear pore complexes (NPCs) requires multiple factors. In Saccharomyces cerevisiae, the NPC proteins Nup159 and Nup42 are asymmetrically localized to the cytoplasmic face and have distinct functional domains: a phenylalanine-glycine (FG) repeat domain that docks mRNP transport receptors and domains that bind the DEAD-box ATPase Dbp5 and its activating cofactor Gle1, respectively. We speculated that the Nup42 and Nup159 FG domains play a role in positioning mRNPs for the terminal mRNP-remodeling steps carried out by Dbp5. Here we find that deletion (Δ) of both the Nup42 and Nup159 FG domains results in a cold-sensitive poly(A)+ mRNA export defect. The nup42ΔFG nup159ΔFG mutant also has synthetic lethal genetic interactions with dbp5 and gle1 mutants. RNA cross-linking experiments further indicate that the nup42ΔFG nup159ΔFG mutant has a reduced capacity for mRNP remodeling during export. To further analyze the role of these FG domains, we replaced the Nup159 or Nup42 FG domains with FG domains from other Nups. These FG "swaps" demonstrate that only certain FG domains are functional at the NPC cytoplasmic face. Strikingly, fusing the Nup42 FG domain to the carboxy-terminus of Gle1 bypasses the need for the endogenous Nup42 FG domain, highlighting the importance of proximal positioning for these factors. We conclude that the Nup42 and Nup159 FG domains target the mRNP to Gle1 and Dbp5 for mRNP remodeling at the NPC. Moreover, these results provide key evidence that character and context play a direct role in FG domain function and mRNA export.
Keywords: DEAD-box protein; mRNA export; nuclear pore complex; nuclear transport; nucleoporin.
Copyright © 2014 by the Genetics Society of America.
Figures
Figure 1
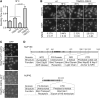
Deletion of FG domains on the cytoplasmic face of the NPC results in a cold-sensitive mRNA export defect. (A) nup42ΔFG nup159ΔFG has a cold-sensitive growth defect. The indicated strains were grown to early log phase (OD600 ∼0.2) at 16°, with OD600 measurements taken every 3 hr and normalized to time = 0, and doubling times were determined. Graph displays average of three independent experiments, and error bars indicate SEM. (B) nup42ΔFG nup159ΔFG has an mRNA export defect at 16°. In situ hybridization with an oligo(dT) probe for poly(A)+ RNA localization was conducted on indicated mutants after growth at 30° and shifting to 16° or 30° overnight. DAPI staining marks the nucleus. Bar, 5 μm. Quantification of three independent experiments of >100 cells for each strain is shown below images. “%N>C” indicates the percentage of cells with increased nuclear poly(A)+ signal (as puncta or diffuse nuclear signal). Uncertainty (±) indicates SEM. (C) nup42ΔFG-GFP and nup159ΔFG-GFP are localized at the NPC. Cells were grown at 30°, shifted to 16° or 30° for 12 hr, and imaged by wide-field live-cell direct fluorescence microscopy. Bar, 5 μm. (D) Diagram depicting functional and structural domains of Nup159 and Nup42 as determined by prior studies (Del Priore et al. 1997; Saavedra et al. 1997; Stutz et al. 1997; Belgareh et al. 1998; Hurwitz et al. 1998; Schmitt et al. 1999; Strahm et al. 1999; Bailer et al. 2000; Vainberg et al. 2000; Denning et al. 2003; Strawn et al. 2004; Weirich et al. 2004; Stelter et al. 2007; Noble et al. 2011; Yoshida et al. 2011) to scale according to primary structure. NTD, amino-terminal domain; DID, dynein-interacting domain; CTD, carboxy-terminal domain.
Figure 2
nup42ΔFG nup159ΔFG exhibits genetic interactions with mRNP-remodeling mutants. (A) nup42ΔFG nup159ΔFG is synthetically lethal with rat8-2 (dbp5). Strains bearing the indicated alleles in addition to a DBP5/URA vector were spotted onto YPD or 5-FOA at 25°. Failure to grow on 5-FOA indicates synthetic lethality. (B) nup42ΔFG is synthetically lethal with gle1-4. Strains bearing the indicated alleles, in addition to a GLE1/URA vector, were spotted onto YPD or 5-FOA at 25°. (C) nup42ΔFG nup159ΔFG is not synthetically lethal with mex67-5. Strains bearing the indicated alleles in addition to a MEX67/URA vector were spotted onto YPD or 5-FOA at 25°. (D) _nup42ΔFG nup159ΔFG gle1_KK>QQ has an enhanced growth defect. Yeast strains were grown at 25° and fivefold serially diluted on YPD plates for growth at the indicated temperature. (E) _nup42ΔFG nup159ΔFG gle1_KK>QQ has an enhanced mRNA export defect. In situ hybridization with an oligo(dT) probe for poly(A)+ RNA localization was conducted on indicated mutants after growth at 25° and shifting to 30° for 3 hr. DAPI staining marks the nucleus. Bar, 5 μm. Quantification of three independent experiments of >100 cells for each strain is shown below images. “%N>C” indicates the percentage of cells with increased nuclear poly(A)+ signal. Uncertainty (±) indicates SEM.
Figure 3
Deletion of the FG domains on the cytoplasmic face of the NPC results in an mRNP remodeling defect in vivo. (A and B) nup42ΔFG nup159ΔFG has increased Nab2 association with poly(A)+ RNA. The association of Nab2 and Cbp80 proteins with poly(A)+ RNA was assessed by shifting strains to 16° overnight, cross-linking with 254-nm UV light, isolating RNA by antisense chromatography, and immunoblotting after treatment with RNase. (A) Representative immunoblot. (B) The level of Nab2 and Cbp80 bound is indicated as a ratio of protein bound to poly(A)+ RNA relative to total cellular protein in each strain, with the wild-type ratio normalized to 1. Graph indicates the average of three independent experiments, and error bars indicate SEM. (C) nab2-C437S partially rescues the cold sensitivity of nup42ΔFG nup159ΔFG. Yeast strains were grown at 30° and fivefold serially diluted on YPD plates for growth at 16°.
Figure 4
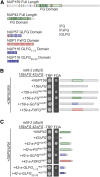
FG swaps reveal specificity of FG domain function. (A) Diagram depicting type and location of FG repeats for indicated FG domains. The FG domain is shown as delineated by Strawn et al. (2001) (for nup116GLFG1-12 and nup116GLFG22-33, see Strawn et al. 2004). Diagram is to scale according to primary structure. (B) The Nup42 FG domain can functionally compensate for the Nup159 FG domain. rat8-2 (dbp5) nup42ΔFG nup159ΔFG mutants containing nup159-s-FG/TRP FG swap vectors in addition to a DBP5/URA vector were spotted onto −Trp synthetic media or 5-FOA at 25°. Growth on 5-FOA indicates functional complementation. (C) The Nup57 GLFG domain and a Nup116 GLFG subdomain that bind Mex67 can functionally compensate for the Nup42 FG domain. rat8-2 (dbp5) nup42ΔFG nup159ΔFG mutants containing nup42-s-FG/TRP FG swap vectors in addition to a DBP5/URA vector were spotted onto −Trp synthetic media or 5-FOA at 25°. Growth on 5-FOA indicates functional complementation.
Figure 5
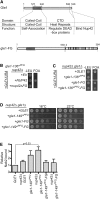
A gle1-FG chimera complements nup42Δ gle1 mutants. (A) Diagram depicting functional and structural domains of Gle1, to scale according to primary structure, and a diagram of the gle1-FG fusion. Asterisk indicates location of PFQ insertions. (B) The Nup42 FG domain is essential when Gle1 self-association is perturbed. nup42Δ gle1-136^PFQ mutants containing a GLE1/URA vector in addition to an empty vector (EV) or nup42/TRP vectors were spotted onto −Trp synthetic media or 5-FOA at 25°. Growth on 5-FOA indicates functional complementation. (C) gle1-136^PFQ-FG rescues synthetic lethality of nup42Δ gle1-136^PFQ. nup42Δ gle1Δ mutants containing a GLE1/URA vector in addition to gle1-136^PFQ/LEU vectors without or with carboxy-terminal FG fusions were spotted onto −Leu synthetic media or 5-FOA at 25°. (D) gle1-149^PFQ-FG partially rescues the synthetic growth defect of nup42Δ gle1-149^PFQ. nup42Δ gle1Δ mutants containing gle1-149^PFQ/LEU vectors without or with carboxy-terminal FG fusions were spotted onto YPD plates to grow at the indicated temperatures. (E) gle1-149^PFQ-FG rescues the mRNP-remodeling defect of nup42Δ gle1-149^PFQ. The association of Nab2 protein with poly(A)+ RNA was assessed by shifting strains to 16° for 3 hr, UV cross-linking, isolating RNA by antisense chromatography, and immunoblotting after treatment with RNase. The level of Nab2 bound is indicated as a ratio of Nab2 bound to poly(A)+ RNA relative to total cellular protein in each strain, with the GLE1 ratio being normalized to 1. Graph indicates the average of three independent experiments, and error bars indicate SEM.
Figure 6
The Nup42 FG domain and CTD have distinct functions. (A) gle1-FG rescues synthetic lethality due to loss of the Nup42 FG domain. rat8-2 (dbp5) nup42ΔFG nup159ΔFG containing empty vector (EV) or gle1/LEU vectors were spotted onto −Leu synthetic media or 5-FOA at 25°. Growth on 5-FOA indicates functional complementation. (B) gle1-FG does not rescue the heat-shock mRNA export defect of nup42Δ mutants. nup42Δ strains containing EV, nup42/LEU, or gle1/LEU vectors were grown at 25° to early log phase, kept at 25° or shifted to 42° for 15 min, labeled with [35S]methionine for an additional 15 min, and lysed. Lysates were separated by SDS-PAGE, and proteins were visualized by autoradiography. The positions of Hsp proteins, induced upon heat shock, are indicated by asterisks. (C) gle1-FG does not rescue temperature sensitivity of nup42Δ ipk1Δ. nup42Δ ipk1Δ containing EV, nup42/LEU, or gle1/LEU vectors were spotted onto −Leu synthetic media at the indicated temperatures.
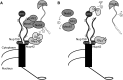
Figure 7
Schematic diagram for model by which FG domains on the cytoplasmic face of the NPC recruit exporting mRNPs for remodeling by Gle1-activated Dbp5. (A) Mex67-Mtr2 mediate export of mature mRNPs, and the dimer is bound to the transcript via adaptors such as the poly(A)+ binding protein Nab2. At the cytoplasmic face of the NPC, the Nup42 and Nup159 FG domains bind Mex67-Mtr2 to bring the mRNP in close proximity to Dbp5. Gle1 stimulates Dbp5 ATP loading, and (i) Dbp5 binds the RNA to trigger remodeling. Dbp5 might also be a constituent of the exporting mRNP. (B) Coincident with ATP hydrolysis, Dbp5 remodels specific proteins such as Nab2 and Mex67 off the mRNP. (ii) These proteins are then recycled into the nucleus for additional rounds of mRNA export. (iii) The mRNP, still bound to other mRNA-binding proteins such as Cbp80, is free for cytoplasmic functions. (iv) The Nup159 NTD facilitates ADP release from Dbp5, allowing additional rounds of mRNP remodeling. It is unknown where the Nup159 NTD is localized relative to its CTD in vivo.
Similar articles
- The Dbp5 cycle at the nuclear pore complex during mRNA export II: nucleotide cycling and mRNP remodeling by Dbp5 are controlled by Nup159 and Gle1.
Noble KN, Tran EJ, Alcázar-Román AR, Hodge CA, Cole CN, Wente SR. Noble KN, et al. Genes Dev. 2011 May 15;25(10):1065-77. doi: 10.1101/gad.2040611. Genes Dev. 2011. PMID: 21576266 Free PMC article. - Nup42 and IP6 coordinate Gle1 stimulation of Dbp5/DDX19B for mRNA export in yeast and human cells.
Adams RL, Mason AC, Glass L, Aditi, Wente SR. Adams RL, et al. Traffic. 2017 Dec;18(12):776-790. doi: 10.1111/tra.12526. Epub 2017 Oct 16. Traffic. 2017. PMID: 28869701 Free PMC article. - The Dbp5 cycle at the nuclear pore complex during mRNA export I: dbp5 mutants with defects in RNA binding and ATP hydrolysis define key steps for Nup159 and Gle1.
Hodge CA, Tran EJ, Noble KN, Alcazar-Roman AR, Ben-Yishay R, Scarcelli JJ, Folkmann AW, Shav-Tal Y, Wente SR, Cole CN. Hodge CA, et al. Genes Dev. 2011 May 15;25(10):1052-64. doi: 10.1101/gad.2041611. Genes Dev. 2011. PMID: 21576265 Free PMC article. - Dbp5, Gle1-IP6 and Nup159: a working model for mRNP export.
Folkmann AW, Noble KN, Cole CN, Wente SR. Folkmann AW, et al. Nucleus. 2011 Nov-Dec;2(6):540-8. doi: 10.4161/nucl.2.6.17881. Epub 2011 Nov 1. Nucleus. 2011. PMID: 22064466 Free PMC article. Review. - Ratcheting mRNA out of the nucleus.
Stewart M. Stewart M. Mol Cell. 2007 Feb 9;25(3):327-30. doi: 10.1016/j.molcel.2007.01.016. Mol Cell. 2007. PMID: 17289581 Review.
Cited by
- Nuclear export of messenger RNA.
Katahira J. Katahira J. Genes (Basel). 2015 Mar 31;6(2):163-84. doi: 10.3390/genes6020163. Genes (Basel). 2015. PMID: 25836925 Free PMC article. Review. - Spelling out the roles of individual nucleoporins in nuclear export of mRNA.
Tingey M, Li Y, Yu W, Young A, Yang W. Tingey M, et al. Nucleus. 2022 Dec;13(1):170-193. doi: 10.1080/19491034.2022.2076965. Nucleus. 2022. PMID: 35593254 Free PMC article. Review. - Of social molecules: The interactive assembly of ASH1 mRNA-transport complexes in yeast.
Niedner A, Edelmann FT, Niessing D. Niedner A, et al. RNA Biol. 2014;11(8):998-1009. doi: 10.4161/rna.29946. Epub 2014 Oct 31. RNA Biol. 2014. PMID: 25482892 Free PMC article. Review. - Structure and Function of the Nuclear Pore Complex Cytoplasmic mRNA Export Platform.
Fernandez-Martinez J, Kim SJ, Shi Y, Upla P, Pellarin R, Gagnon M, Chemmama IE, Wang J, Nudelman I, Zhang W, Williams R, Rice WJ, Stokes DL, Zenklusen D, Chait BT, Sali A, Rout MP. Fernandez-Martinez J, et al. Cell. 2016 Nov 17;167(5):1215-1228.e25. doi: 10.1016/j.cell.2016.10.028. Epub 2016 Nov 10. Cell. 2016. PMID: 27839866 Free PMC article. - Cytoplasmic hGle1A regulates stress granules by modulation of translation.
Aditi, Folkmann AW, Wente SR. Aditi, et al. Mol Biol Cell. 2015 Apr 15;26(8):1476-90. doi: 10.1091/mbc.E14-11-1523. Epub 2015 Feb 18. Mol Biol Cell. 2015. PMID: 25694449 Free PMC article.
References
- Alber F., Dokudovskaya S., Veenhoff L. M., Zhang W., Kipper J., et al. , 2007. The molecular architecture of the nuclear pore complex. Nature 450: 695–701. - PubMed
- Alcazar-Roman A. R., Tran E. J., Guo S., Wente S. R., 2006. Inositol hexakisphosphate and Gle1 activate the DEAD-box protein Dbp5 for nuclear mRNA export. Nat. Cell Biol. 8: 711–716. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- T32 HD007502/HD/NICHD NIH HHS/United States
- R01 GM051219/GM/NIGMS NIH HHS/United States
- R37 GM051219/GM/NIGMS NIH HHS/United States
- T32HD007502/HD/NICHD NIH HHS/United States
- R37GM051219/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases