Human intestinal tissue with adult stem cell properties derived from pluripotent stem cells - PubMed (original) (raw)
. 2014 Jun 3;2(6):838-52.
doi: 10.1016/j.stemcr.2014.05.001.
Kunitoshi Chiba 1, Lorian Schaeffer 1, Samuel G Regalado 1, Christine S Lai 2, Qing Gao 2, Samira Kiani 2, Henner F Farin 3, Hans Clevers 4, Gregory J Cost 5, Andy Chan 5, Edward J Rebar 5, Fyodor D Urnov 5, Philip D Gregory 5, Lior Pachter 6, Rudolf Jaenisch 7, Dirk Hockemeyer 1
Affiliations
- PMID: 24936470
- PMCID: PMC4050346
- DOI: 10.1016/j.stemcr.2014.05.001
Human intestinal tissue with adult stem cell properties derived from pluripotent stem cells
Ryan Forster et al. Stem Cell Reports. 2014.
Erratum in
- Stem Cell Reports. 2014 Jul 8;3(1):215
Abstract
Genetically engineered human pluripotent stem cells (hPSCs) have been proposed as a source for transplantation therapies and are rapidly becoming valuable tools for human disease modeling. However, many applications are limited due to the lack of robust differentiation paradigms that allow for the isolation of defined functional tissues. Here, using an endogenous LGR5-GFP reporter, we derived adult stem cells from hPSCs that gave rise to functional human intestinal tissue comprising all major cell types of the intestine. Histological and functional analyses revealed that such human organoid cultures could be derived with high purity and with a composition and morphology similar to those of cultures obtained from human biopsies. Importantly, hPSC-derived organoids responded to the canonical signaling pathways that control self-renewal and differentiation in the adult human intestinal stem cell compartment. This adult stem cell system provides a platform for studying human intestinal disease in vitro using genetically engineered hPSCs.
Figures
Figure 1
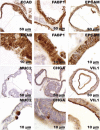
Generation of LGR5-GFP Reporter hESCs Using ZFNs (A) Schematic overview depicting the gene-editing strategy for the LGR5 locus using a ZFN targeted to either the first or last coding exon of LGR5. Southern blot probes are shown as red boxes, exons are shown as blue boxes. Below are the donor plasmids used to target the LGR5 locus to generate either an N-terminal (LGR5-GFPN-term) or a C-terminal (LGR5-GFPC-term) LGR5-GFP reporter. pA, polyadenylation sequence; PGK, phosphoglycerate kinase promoter; Puro, puromycin resistance gene; eGFP, enhanced green fluorescent protein. Shown below the donor plasmids is the LGR5 locus after targeting with the respective donor plasmids. (B) IHC staining for indicated proteins in teratoma sections derived from LGR5-GFPN-term hESC reporter cells. MUC2, mucin2; VIL1, villin1; CHGA, chromogranin A; CDX2, caudal type homeobox 2; LYZ, lysozyme; PDX1, pancreatic and duodenal homeobox 1. (C) Phase contrast and H&E staining of epithelial organoid cultures arising from LGR5-GFP sorting experiments. See quantification in Table 1. (D) Bright-field images of developing organoids (outlined by yellow lines) derived from nonsorted single-cell suspensions after Matrigel embedding. The demarcation outlined by white lines in bright-field images taken on days 5∗–15 was used as a reference point for tracking organoids clustered near this region. Cells were passaged every 5 days; here, an image before and after (∗) passaging is shown. All bars, 200 μm. See also Figure S1.
Figure 2
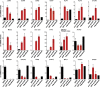
Isolation of Intestinal Organoids Independent of the LGR5-GFP Reporter System Expression profiling of organoids from GFP-positive and nonsorted LGR5-GFP teratoma cells compared with hESCs and fibroblast-like cells. qRT-PCR for the indicated genes in hESCs, organoids derived from eGFP-positive cells (n = 3), and nonsorted (n = 3) cells of LGR5-GFPN-term, hESCs, and fibroblast-like cells (derived from hESCs as described previously [Hockemeyer et al., 2008], expressing telomerase from the AAVS1 locus). Relative expression levels were normalized to baseline expression of these genes in hESCs. Data are biological replicates of independent experiments; bars represent the SEM. See also Figure S2.
Figure 3
hESC-Derived Organoids Comprise Specific Cell Types Characteristic of the Human Intestinal Epithelium (A) IHC staining for intestinal marker proteins in sections of hESC-derived organoids generated from wild-type hESC reporter cells. ECAD1, E-cadherin; FABP1, fatty acid-binding protein 1; EPCAM, epithelial cell adhesion molecule; MUC2, mucin2; CHGA, chromogranin A; VIL1, villin1. See also Figure S3.
Figure 4
Subcellular Organization of Organoids Resembles the Structure of the Human Intestinal Epithelium (A) Electron micrograph of hESC-derived organoids. Shown is a representative image with enterocyte-like cells forming a polarized epithelium. The lumen of the organoid is oriented to the left (apical), while the nuclei are aligned along the right (basal). (B) Higher-magnification images of the micrograph shown in (A). Insets indicate areas of increased magnification. Images show the microvilli lining the luminal cell surface, vesicles in the luminal space (II), and the tight junctions connecting adjacent enterocyte-like cells (I). (C) Electron micrograph of hESC-derived organoids. Shown is a representative image of a goblet-like secretory cell that is embedded into a sheet of enterocyte-like cells. Orientation of the image as in (A). (D) Higher-magnification images of the micrograph shown in (C). Insets indicate areas of increased magnification. Images show magnification of vesicles (I), rough endoplasmic reticulum at the base of the vesicles (II), and tight junctions and cellular debris shed into the organoid’s lumen (III).
Figure 5
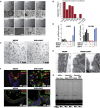
WNT and Notch Signaling Is Required to Establish and Maintain Cells with Adult Stem Cell Properties in hESC-Derived Intestinal Organoids (A) Bright-field image of organoids at day 5 of their derivation under the indicated culture conditions at two different magnifications. Growth factors (GFs) : WENR [W, Wnt3a; E, EGF; N, Noggin; R, R-spondin-1] were supplemented in the combinations indicated. No GFs were added to “base media.” (B) Quantification of images shown in (A). The graph shows the number of organoids larger than 150 μm from a single teratoma isolation cultured in parallel under the conditions described in (A), where organoids were grown in 50 μl solidified Matrigel in 500 μl media/well in a 24-well plate. (C) Bright-field image of organoids at day 15. The image to the left shows a culture at day 15 when grown in WENR, and the right image shows the same cells when switched at day 10 to differentiation media. (D) qRT-PCR for the intestinal stem cell markers LGR5 and OLFM4 in a single organoid culture treated in parallel with different culture conditions. The ‡ symbol indicates 1,000 ng/ml and 200 ng/ml of RSPO1 and WNT3a, respectively, and + indicates 200 μg/ml and 50 ng/ml of RSPO1 and WNT3a, respectively. Where indicated, DAPT was added at 10 μM. (E) Electron micrographs of the cells shown in (C). Size bars indicate 2 μm. (F) IF staining of cryosectioned organoids cultured in either stem cell media (WENR) or differentiation media (10 μM ENR+DAPT added for 4 consecutive days). Top: MUC2 (green), phalloidin (red), and DAPI (blue). Bottom: CDX2 (red), phalloidin (green), and DAPI (blue). (G) TRAP assay (Kim and Wu, 1997) of hESCs, hESCs differentiated into fibroblast-like cells (Hockemeyer et al., 2008), intestinal organoids grown in stem cell media (WENR, n = 2) or differentiation media (10 μm ENR+ DAPT, n = 2). Shown is a 32P autoradiogram of TRAP activity described for decreasing amounts of protein extracts (1.0–0.4 μg) for the indicated cell types (hESCs [WIBR3]; Fibro., fibroblast-like cells derived from WIBR3). See also Figure S4.
Figure 6
hESC-Derived Organoids Share a Transcriptional Profile with Primary Intestinal Tissue-Derived Organoids and Display the Characteristic Responses to Differentiation Stimuli (A) Cluster analysis of hESCs (n = 4), teratoma samples (n = 4), bulk hESC-derived organoid cultures grown in WENR (n = 10), bulk cultures of hESC-derived organoids grown in differentiation medium (ENR+DAPT, n = 4), and primary duodenum- (n = 2), rectum- (n = 2) and ileum- (n = 2) derived organoids. Euclidian distances calculated from the abundance levels of the top 5,000 differentially expressed transcripts are also shown in Figure S1A. (B) Heatmap displaying an unbiased cluster analysis of samples analyzed in (A) for a limited selection of genes relevant to intestinal expression and, in most cases, reported functions. Shown are the Euclidian distances calculated from the relative expression of the genes as determined by next-generation RNA-seq expression analysis. (C) Density maps of RNA-seq reads mapped to the genetically engineered LGR5-GFPN-term and LGR5-GFPC-term locus. The top boxes show the predicted transcripts for each allele based on validated gene editing described in Figure 1A. Shaded in green is the region mapping to the coding sequence for the eGPF fusion reporter. Shown across each histogram are the alignments to Lgr5-eGFP fusions predicted for each cell line. RNA was collected from the parent hESC cell lines, the intermediate teratoma samples, intestinal organoids, and intestinal organoids differentiated by the withdrawal of WNT3a and the addition of DAPT. (D) Gene Ontology analysis for tissue-specific expression using the Database for Annotation, Visualization and Integrated Discovery (DAVID;
http://david.abcc.ncifcrf.gov/home.jsp
) and the “UNIGENE_EST_QUARTILE” expression profile database. Analyzed were genes significantly (FDR corrected p value < 0.05) unregulated (>22.5-fold) in hESC-derived intestinal organoids (n = 10) compared with hESCs (n = 4). (E) Heatmap displaying an unbiased cluster analysis of organoid samples (from groups 1, 2, 6, and 7 as described in Table S1) analyzed in (A) for selected genes that are associated with the differentiation of adult intestinal stem cells. Shown are pairwise hESC-derived intestinal organoid cultures (n = 4) grown in either WENR stem cell media (S) or differentiated in ENR+DAPT (D). See also Figure S5.
Similar articles
- Intestinal Commitment and Maturation of Human Pluripotent Stem Cells Is Independent of Exogenous FGF4 and R-spondin1.
Tamminen K, Balboa D, Toivonen S, Pakarinen MP, Wiener Z, Alitalo K, Otonkoski T. Tamminen K, et al. PLoS One. 2015 Jul 31;10(7):e0134551. doi: 10.1371/journal.pone.0134551. eCollection 2015. PLoS One. 2015. PMID: 26230325 Free PMC article. - Establishment of 3D Intestinal Organoid Cultures from Intestinal Stem Cells.
Sugimoto S, Sato T. Sugimoto S, et al. Methods Mol Biol. 2017;1612:97-105. doi: 10.1007/978-1-4939-7021-6_7. Methods Mol Biol. 2017. PMID: 28634937 - Isolation and Culture of Adult Intestinal, Gastric, and Liver Organoids for Cre-recombinase-Mediated Gene Deletion.
Flanagan DJ, Schwab RHM, Tran BM, Phesse TJ, Vincan E. Flanagan DJ, et al. Methods Mol Biol. 2019;1576:123-133. doi: 10.1007/7651_2016_14. Methods Mol Biol. 2019. PMID: 27704362 - A critical look: Challenges in differentiating human pluripotent stem cells into desired cell types and organoids.
Fowler JL, Ang LT, Loh KM. Fowler JL, et al. Wiley Interdiscip Rev Dev Biol. 2020 May;9(3):e368. doi: 10.1002/wdev.368. Epub 2019 Nov 19. Wiley Interdiscip Rev Dev Biol. 2020. PMID: 31746148 Free PMC article. Review. - Human Enteroids/Colonoids and Intestinal Organoids Functionally Recapitulate Normal Intestinal Physiology and Pathophysiology.
Zachos NC, Kovbasnjuk O, Foulke-Abel J, In J, Blutt SE, de Jonge HR, Estes MK, Donowitz M. Zachos NC, et al. J Biol Chem. 2016 Feb 19;291(8):3759-66. doi: 10.1074/jbc.R114.635995. Epub 2015 Dec 16. J Biol Chem. 2016. PMID: 26677228 Free PMC article. Review.
Cited by
- Evaluating Strategies to Assess the Differentiation Potential of Human Pluripotent Stem Cells: A Review, Analysis and Call for Innovation.
Smith L, Quelch-Cliffe R, Liu F, Aguilar AH, Przyborski S. Smith L, et al. Stem Cell Rev Rep. 2024 Sep 28. doi: 10.1007/s12015-024-10793-5. Online ahead of print. Stem Cell Rev Rep. 2024. PMID: 39340737 Review. - Investigating nanoplastics toxicity using advanced stem cell-based intestinal and lung in vitro models.
Busch M, Brouwer H, Aalderink G, Bredeck G, Kämpfer AAM, Schins RPF, Bouwmeester H. Busch M, et al. Front Toxicol. 2023 Jan 27;5:1112212. doi: 10.3389/ftox.2023.1112212. eCollection 2023. Front Toxicol. 2023. PMID: 36777263 Free PMC article. Review. - Nanoparticles-mediated CRISPR-Cas9 gene therapy in inherited retinal diseases: applications, challenges, and emerging opportunities.
Chien Y, Hsiao YJ, Chou SJ, Lin TY, Yarmishyn AA, Lai WY, Lee MS, Lin YY, Lin TW, Hwang DK, Lin TC, Chiou SH, Chen SJ, Yang YP. Chien Y, et al. J Nanobiotechnology. 2022 Dec 3;20(1):511. doi: 10.1186/s12951-022-01717-x. J Nanobiotechnology. 2022. PMID: 36463195 Free PMC article. Review. - Organoids in gastrointestinal diseases: from experimental models to clinical translation.
Günther C, Winner B, Neurath MF, Stappenbeck TS. Günther C, et al. Gut. 2022 Sep;71(9):1892-1908. doi: 10.1136/gutjnl-2021-326560. Epub 2022 May 30. Gut. 2022. PMID: 35636923 Free PMC article. Review. - Engineering multiscale structural orders for high-fidelity embryoids and organoids.
Shao Y, Fu J. Shao Y, et al. Cell Stem Cell. 2022 May 5;29(5):722-743. doi: 10.1016/j.stem.2022.04.003. Cell Stem Cell. 2022. PMID: 35523138 Free PMC article. Review.
References
- Barker N., van Es J.H., Kuipers J., Kujala P., van den Born M., Cozijnsen M., Haegebarth A., Korving J., Begthel H., Peters P.J., Clevers H. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature. 2007;449:1003–1007. - PubMed
- Cao L., Gibson J.D., Miyamoto S., Sail V., Verma R., Rosenberg D.W., Nelson C.E., Giardina C. Intestinal lineage commitment of embryonic stem cells. Differentiation. 2011;81:1–10. - PubMed
- de Lau W., Barker N., Low T.Y., Koo B.-K., Li V.S.W., Teunissen H., Kujala P., Haegebarth A., Peters P.J., van de Wetering M. Lgr5 homologues associate with Wnt receptors and mediate R-spondin signalling. Nature. 2011;476:293–297. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- T32 GM007232/GM/NIGMS NIH HHS/United States
- R37-CA084198/CA/NCI NIH HHS/United States
- R37 CA084198/CA/NCI NIH HHS/United States
- R01 HD045022/HD/NICHD NIH HHS/United States
- R01 CA087869/CA/NCI NIH HHS/United States
- 2T32GM007232-36/GM/NIGMS NIH HHS/United States
- T32 GM066698/GM/NIGMS NIH HHS/United States
- R01-CA087869/CA/NCI NIH HHS/United States
- P30 CA014051/CA/NCI NIH HHS/United States
- R01-HD045022/HD/NICHD NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources