Prognostic value of isocitrate dehydrogenase mutations in myelodysplastic syndromes: a retrospective cohort study and meta-analysis - PubMed (original) (raw)
Meta-Analysis
Prognostic value of isocitrate dehydrogenase mutations in myelodysplastic syndromes: a retrospective cohort study and meta-analysis
Jie Jin et al. PLoS One. 2014.
Abstract
Background: Recent genomic sequencing efforts have identified a number of recurrent mutations in myelodysplastic syndromes (MDS) that may contribute to disease progression and overall survival, including mutations in isocitrate dehydrogenases 1 and 2 (IDH1 and IDH2).
Methods: Pretreatment bone marrow (BM) samples were acquired from mononuclear cells in 146 adult patients with de novo MDS from January 2006 to June 2013. Polymerase chain reaction (PCR) and direct sequencing were performed on exon 4 of IDH1/2 genes and mutation status was correlated with overall survival (OS) and leukemia-free survival (LFS). We then performed a meta-analysis combining previously published and current studies to explore the effect of IDH mutations on OS and LFS in MDS.
Results: In our study, somatic mutations of either IDH gene were discovered in 11 MDS patients (7.53%) and were significantly correlated with poorer OS (P = 0.007). IDH mutations were specifically associated with a poorer OS in the intermediate-1 risk group by the International Prognostic Scoring System (IPSS) (P = 0.039). In addition, we discovered decitabine achieved a better therapeutic effect compared to other treatments in IDH mutation-positive patients (P = 0.023). We identified six previous studies of IDH mutations in MDS. A meta-analysis of these studies included 111 MDS patients IDH mutations and 1671 MDS patients with wild-type IDH1/2. The hazard ratios (HRs) of OS and LFS for patients with IDH mutations were 1.62 (95% CI, 1.27-2.09) and 2.21 (95% CI, 1.48-3.30), respectively.
Conclusion: The results from our study and the meta-analysis provide firm evidence that IDH mutations are significantly associated with poorer clinical outcomes in MDS. Identification of IDH mutations may be pivotal for better risk stratification in MDS patients and improving IPSS score. Additionally, hypomethylating agents may be an effective treatment option for MDS patients with IDH mutations.
Conflict of interest statement
Competing Interests: The authors have declared that no competing interests exist.
Figures
Figure 1. Flow diagram of study selection.
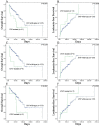
Figure 2. Kaplan–Meier survival curves for survival of MDS patients.
(A) Overall survival data for MDS patients stratified by IDH1/2 mutational status. (B) Leukemia-free survival data for MDS patients stratified by IDH1/2 mutational status. (C) Overall survival data for MDS patients stratified by IDH1 mutational status. (D) Leukemia-free survival data for MDS patients stratified by IDH1 mutational status. (E) Overall survival data for MDS patients stratified by IDH2 mutational status. (F) Leukemia-free survival data for MDS patients stratified by IDH2 mutational status.
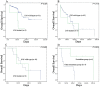
Figure 3. Kaplan–Meier survival curves for overall survival of MDS patients.
(A) Overall survival of MDS patients in the intermediate-1 risk group of IPSS. (B) Overall survival of MDS patients in the intermediate-2 risk group of IPSS. (C) Overall survival of MDS patients in the high risk group of IPSS. (D) Kaplan–Meier survival of IDH mutant patients with decitabine chemotherapy compared with other treatments.
Figure 4. Forest plots describing the association between IDH mutations and MDS.
(A) Forest plots of HR and 95% CI for IDH1/2 mutations in MDS comparing with IDH wild-type by OS endpoints. (B) Forest plots of HR and 95% CI for IDH1/2 mutations in MDS comparing with IDH wild-type by LFS endpoints. (C) Forest plots of HR and 95% CI for _IDH_1 mutations in MDS comparing with _IDH_1 wild-type by OS endpoints. (D) Forest plots of HR and 95% CI for _IDH_1 mutations in MDS comparing with _IDH_1 wild-type by LFS endpoints. (E) Forest plots of HR and 95% CI for _IDH_2 mutations in MDS comparing with _IDH_2 wild-type by OS endpoints. (F) Forest plots of cumulative meta-analysis of IDH mutations in association with MDS for OS by published year.
Similar articles
- Isocitrate dehydrogenase mutations in myeloid malignancies.
Medeiros BC, Fathi AT, DiNardo CD, Pollyea DA, Chan SM, Swords R. Medeiros BC, et al. Leukemia. 2017 Feb;31(2):272-281. doi: 10.1038/leu.2016.275. Epub 2016 Oct 10. Leukemia. 2017. PMID: 27721426 Free PMC article. Review. - [Relationship between clinical characteristics and myelodysplastic syndrome patients with isocitrate dehydrogenase gene mutations].
Tong HY, Hu C, Yu MX, Ma QL, Chen FF, Ye L, Wei JY, Xu GX, Mao LP, Li Y, Jin J. Tong HY, et al. Zhonghua Yi Xue Za Zhi. 2013 Oct 29;93(40):3180-4. Zhonghua Yi Xue Za Zhi. 2013. PMID: 24405536 Chinese. - IDH1 Mutation Is an Independent Inferior Prognostic Indicator for Patients with Myelodysplastic Syndromes.
Wang N, Wang F, Shan N, Sui X, Xu H. Wang N, et al. Acta Haematol. 2017;138(3):143-151. doi: 10.1159/000479546. Epub 2017 Sep 6. Acta Haematol. 2017. PMID: 28873367 - Isocitrate dehydrogenase 2 mutations correlate with leukemic transformation and are predicted by 2-hydroxyglutarate in myelodysplastic syndromes.
Lin P, Luo Y, Zhu S, Maggio D, Yang H, Hu C, Wang J, Zhang H, Ren Y, Zhou X, Mei C, Ma L, Xu W, Ye L, Zhuang Z, Jin J, Tong H. Lin P, et al. J Cancer Res Clin Oncol. 2018 Jun;144(6):1037-1047. doi: 10.1007/s00432-018-2627-3. Epub 2018 Mar 16. J Cancer Res Clin Oncol. 2018. PMID: 29549529 - Prognostic significance of SRSF2 mutations in myelodysplastic syndromes and chronic myelomonocytic leukemia: a meta-analysis.
Arbab Jafari P, Ayatollahi H, Sadeghi R, Sheikhi M, Asghari A. Arbab Jafari P, et al. Hematology. 2018 Dec;23(10):778-784. doi: 10.1080/10245332.2018.1471794. Epub 2018 May 14. Hematology. 2018. PMID: 29757120 Review.
Cited by
- Lower-dose decitabine improves clinical response compared with best supportive care in lower-risk MDS patients: a prospective, multicenter phase 2 study.
Ye L, Mei C, Ren Y, Zhou X, Ma L, Xu W, Wei J, Jiang H, Zhang L, Zeng H, Tong H. Ye L, et al. J Cancer. 2021 Mar 19;12(10):2975-2981. doi: 10.7150/jca.56207. eCollection 2021. J Cancer. 2021. PMID: 33854598 Free PMC article. - Hypomethylating agent combination strategies in myelodysplastic syndromes: hopes and shortcomings.
Ball B, Zeidan A, Gore SD, Prebet T. Ball B, et al. Leuk Lymphoma. 2017 May;58(5):1022-1036. doi: 10.1080/10428194.2016.1228927. Epub 2016 Sep 21. Leuk Lymphoma. 2017. PMID: 27654579 Free PMC article. Review. - The spectrum of genetic mutations in myelodysplastic syndrome: Should we update prognostication?
Cook MR, Karp JE, Lai C. Cook MR, et al. EJHaem. 2021 Nov 1;3(1):301-313. doi: 10.1002/jha2.317. eCollection 2022 Feb. EJHaem. 2021. PMID: 35846202 Free PMC article. Review. - Beyond the Edge of Hypomethylating Agents: Novel Combination Strategies for Older Adults with Advanced MDS and AML.
Kubasch AS, Platzbecker U. Kubasch AS, et al. Cancers (Basel). 2018 May 24;10(6):158. doi: 10.3390/cancers10060158. Cancers (Basel). 2018. PMID: 29795051 Free PMC article. Review. - Isocitrate dehydrogenase mutations in myeloid malignancies.
Medeiros BC, Fathi AT, DiNardo CD, Pollyea DA, Chan SM, Swords R. Medeiros BC, et al. Leukemia. 2017 Feb;31(2):272-281. doi: 10.1038/leu.2016.275. Epub 2016 Oct 10. Leukemia. 2017. PMID: 27721426 Free PMC article. Review.
References
- Corey SJ, Minden MD, Barber DL, Kantarjian H, Wang JC, et al. (2007) Myelodysplastic syndromes: the complexity of stem-cell diseases. Nat Rev Cancer 7: 118–129. - PubMed
- Nimer SD (2008) Myelodysplastic syndromes. Blood 111: 4841–4851. - PubMed
- Greenberg P, Cox C, LeBeau MM, Fenaux P, Morel P, et al. (1997) International scoring system for evaluating prognosis in myelodysplastic syndromes. Blood 89: 2079–2088. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
This study was supported by grants from Zhejiang Province Fund for Distinguished Young Scholars (LR12H08001), the Foundation of Key Innovation Team of Zhejiang Province (2011R50015), National Public Health Grand Research Foundation (201202017), major program of Science Technology Department of Zhejiang Province fund (2013c03043-2) and the National Natural Science Foundation of China (No.30870914, No.81270582). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous