FOCUS: an alignment-free model to identify organisms in metagenomes using non-negative least squares - PubMed (original) (raw)
FOCUS: an alignment-free model to identify organisms in metagenomes using non-negative least squares
Genivaldo Gueiros Z Silva et al. PeerJ. 2014.
Abstract
One of the major goals in metagenomics is to identify the organisms present in a microbial community from unannotated shotgun sequencing reads. Taxonomic profiling has valuable applications in biological and medical research, including disease diagnostics. Most currently available approaches do not scale well with increasing data volumes, which is important because both the number and lengths of the reads provided by sequencing platforms keep increasing. Here we introduce FOCUS, an agile composition based approach using non-negative least squares (NNLS) to report the organisms present in metagenomic samples and profile their abundances. FOCUS was tested with simulated and real metagenomes, and the results show that our approach accurately predicts the organisms present in microbial communities. FOCUS was implemented in Python. The source code and web-sever are freely available at http://edwards.sdsu.edu/FOCUS.
Keywords: Metagenomes; Modeling; k-mer.
Figures
Figure 1. Workflow of the FOCUS program.
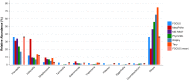
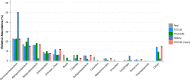
Figure 2. Genera-level taxonomy classification sorted by FOCUS prediction for the metagenome from a diseased human oral cavity using FOCUS, MetaPhlAn, MG-RAST, PhymnBL, RAIphy, Taxy, and FOCUS (mean).
Error bars represent the standard deviation uncertainty in tested metagenome.
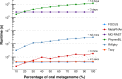
Figure 3. Scalability test using different sub-sets of the human oral cavity under disease metagenome using FOCUS, MetaPhlAn, MG-RAST, PhymnBL, RAIphy, Taxy.
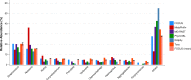
Figure 4. Genera-level taxonomy classification sorted by FOCUS prediction for the metagenome from a healthy human oral cavity using FOCUS, MetaPhlAn, MG-RAST, PhymnBL, RAIphy, Taxy, and FOCUS (mean).
Error bars show the standard deviation for the real metagenome.
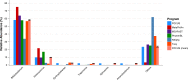
Figure 5. Genera-level taxonomy classification sorted by FOCUS prediction for the metagenome from a fecal metagenomic sample of a healthy human using FOCUS, MetaPhlAn, MG-RAST, PhymnBL, RAIphy, Taxy, and FOCUS (mean).
Error bars show the standard deviation for the real metagenome.
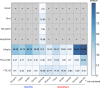
Figure 6. Heat-map representing the distance between the FOCUS and MetaPhlAn results for 300 metagenomes from the Human Microbiome Project across 15 body sites.
The distance was computed using the Euclidean distance between the results of both tools.
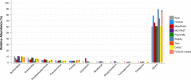
Figure 7. Genera-level taxonomy classification for the SimShort dataset using FOCUS, PhymnBL, RAIphy, and FOCUS (mean).
Figure 8. Class-level taxonomy classification for the SimHC dataset using FOCUS, PhymnBL, RAIphy, and FOCUS (mean).
Figure 9. Genera-level taxonomy classification for the SimHC dataset using FOCUS, MetaPhlAn, MG-RAST, PhymnBL, RAIphy, Taxy, GASiC, and FOCUS (mean).
Figure 10. Numerical evaluation between the real and predicted abundance for the synthetic metagenomes computed by the Euclidean distance between the real and the predicted values.
Similar articles
- SUPER-FOCUS: a tool for agile functional analysis of shotgun metagenomic data.
Silva GG, Green KT, Dutilh BE, Edwards RA. Silva GG, et al. Bioinformatics. 2016 Feb 1;32(3):354-61. doi: 10.1093/bioinformatics/btv584. Epub 2015 Oct 9. Bioinformatics. 2016. PMID: 26454280 Free PMC article. - An Agile Functional Analysis of Metagenomic Data Using SUPER-FOCUS.
Silva GGZ, Lopes FAC, Edwards RA. Silva GGZ, et al. Methods Mol Biol. 2017;1611:35-44. doi: 10.1007/978-1-4939-7015-5_4. Methods Mol Biol. 2017. PMID: 28451970 - Assessment of k-mer spectrum applicability for metagenomic dissimilarity analysis.
Dubinkina VB, Ischenko DS, Ulyantsev VI, Tyakht AV, Alexeev DG. Dubinkina VB, et al. BMC Bioinformatics. 2016 Jan 16;17:38. doi: 10.1186/s12859-015-0875-7. BMC Bioinformatics. 2016. PMID: 26774270 Free PMC article. - Assessment of metagenomic assemblers based on hybrid reads of real and simulated metagenomic sequences.
Wang Z, Wang Y, Fuhrman JA, Sun F, Zhu S. Wang Z, et al. Brief Bioinform. 2020 May 21;21(3):777-790. doi: 10.1093/bib/bbz025. Brief Bioinform. 2020. PMID: 30860572 Free PMC article. Review. - What Is Metagenomics Teaching Us, and What Is Missed?
New FN, Brito IL. New FN, et al. Annu Rev Microbiol. 2020 Sep 8;74:117-135. doi: 10.1146/annurev-micro-012520-072314. Epub 2020 Jun 30. Annu Rev Microbiol. 2020. PMID: 32603623 Review.
Cited by
- Multidimensional metrics for estimating phage abundance, distribution, gene density, and sequence coverage in metagenomes.
Aziz RK, Dwivedi B, Akhter S, Breitbart M, Edwards RA. Aziz RK, et al. Front Microbiol. 2015 May 8;6:381. doi: 10.3389/fmicb.2015.00381. eCollection 2015. Front Microbiol. 2015. PMID: 26005436 Free PMC article. - Metagenomic Functional Potential Predicts Degradation Rates of a Model Organophosphorus Xenobiotic in Pesticide Contaminated Soils.
Jeffries TC, Rayu S, Nielsen UN, Lai K, Ijaz A, Nazaries L, Singh BK. Jeffries TC, et al. Front Microbiol. 2018 Feb 20;9:147. doi: 10.3389/fmicb.2018.00147. eCollection 2018. Front Microbiol. 2018. PMID: 29515526 Free PMC article. - Contrasting the Genetic Patterns of Microbial Communities in Soda Lakes with and without Cyanobacterial Bloom.
Andreote APD, Dini-Andreote F, Rigonato J, Machineski GS, Souza BCE, Barbiero L, Rezende-Filho AT, Fiore MF. Andreote APD, et al. Front Microbiol. 2018 Feb 22;9:244. doi: 10.3389/fmicb.2018.00244. eCollection 2018. Front Microbiol. 2018. PMID: 29520256 Free PMC article. - PhyloPythiaS+: a self-training method for the rapid reconstruction of low-ranking taxonomic bins from metagenomes.
Gregor I, Dröge J, Schirmer M, Quince C, McHardy AC. Gregor I, et al. PeerJ. 2016 Feb 8;4:e1603. doi: 10.7717/peerj.1603. eCollection 2016. PeerJ. 2016. PMID: 26870609 Free PMC article. - Environmental genes and genomes: understanding the differences and challenges in the approaches and software for their analyses.
Zepeda Mendoza ML, Sicheritz-Pontén T, Gilbert MT. Zepeda Mendoza ML, et al. Brief Bioinform. 2015 Sep;16(5):745-58. doi: 10.1093/bib/bbv001. Epub 2015 Feb 11. Brief Bioinform. 2015. PMID: 25673291 Free PMC article.
References
- Aziz RK, Devoid S, Disz T, Edwards RA, Henry CS, Olsen GJ, Olson R, Overbeek R, Parrello B, Pusch GD, Stevens RL, Vonstein V, Xia F. SEED servers: high-performance access to the seed genomes, annotations, and metabolic models. PLoS ONE. 2012;7:e425. doi: 10.1371/journal.pone.0048053. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous