Altered translation of GATA1 in Diamond-Blackfan anemia - PubMed (original) (raw)
doi: 10.1038/nm.3557. Epub 2014 Jun 22.
Hanna T Gazda 2, Jennifer C Eng 3, Stephen W Eichhorn 4, Prathapan Thiru 5, Roxanne Ghazvinian 6, Tracy I George 7, Jason R Gotlib 8, Alan H Beggs 9, Colin A Sieff 10, Harvey F Lodish 11, Eric S Lander 12, Vijay G Sankaran 13
Affiliations
- PMID: 24952648
- PMCID: PMC4087046
- DOI: 10.1038/nm.3557
Altered translation of GATA1 in Diamond-Blackfan anemia
Leif S Ludwig et al. Nat Med. 2014 Jul.
Abstract
Ribosomal protein haploinsufficiency occurs in diverse human diseases including Diamond-Blackfan anemia (DBA), congenital asplenia and T cell leukemia. Yet, how mutations in genes encoding ubiquitously expressed proteins such as these result in cell-type- and tissue-specific defects remains unknown. Here, we identify mutations in GATA1, encoding the critical hematopoietic transcription factor GATA-binding protein-1, that reduce levels of full-length GATA1 protein and cause DBA in rare instances. We show that ribosomal protein haploinsufficiency, the more common cause of DBA, can lead to decreased GATA1 mRNA translation, possibly resulting from a higher threshold for initiation of translation of this mRNA in comparison with other mRNAs. In primary hematopoietic cells from patients with mutations in RPS19, encoding ribosomal protein S19, the amplitude of a transcriptional signature of GATA1 target genes was globally and specifically reduced, indicating that the activity, but not the mRNA level, of GATA1 is decreased in patients with DBA associated with mutations affecting ribosomal proteins. Moreover, the defective hematopoiesis observed in patients with DBA associated with ribosomal protein haploinsufficiency could be partially overcome by increasing GATA1 protein levels. Our results provide a paradigm by which selective defects in translation due to mutations affecting ubiquitous ribosomal proteins can result in human disease.
Conflict of interest statement
The authors declare no competing financial interests.
Figures
Figure 1. GATA1 full-length protein production is necessary for human erythropoiesis and its disruption results in DBA
(a) Sanger sequencing of the first initiator codon region from exon 2 of GATA1 in a male DBA patient, the patient’s mother, and a healthy control. (b) Bone marrow aspirate (above at 100X objective magnification with scale bar at bottom right showing 10 μM distance) and biopsy (below at 50X objective magnification with scale bar showing 30 μM distance) sections from marrow of the DBA patient. (c) The production of full-length and short protein forms of GATA1 from the full-length mRNA with exogenous expression of either no vector, HMD lentiviral vector, HMD-GATA1 wildtype, or the HMD-GATA1 mutant vectors in 293T cells. Arrowheads indicate the major protein forms of GATA1 full length and GATA1 short. (d) Western blots against GATA1 (C-terminal antibody) with a β-actin loading control. Arrowheads indicate the major protein forms of GATA1 full length and GATA1 short.
Figure 2. Ribosomal protein haploinsufficiency results in reduced translation of GATA1
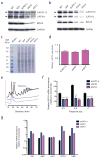
(a) Protein lysates from CD34+ derived erythroid progenitors at the CFU-E to proerythroblast stage (day 4 of differentiation) from cells infected with RPS19 shRNA vectors, sh916 and sh913. Arrowheads indicate the major protein forms of GATA1 full length and GATA1 short. (b) Protein lysates from K562 cells infected with RPS19 shRNA vectors, sh916 and sh913. Arrowheads indicate the major protein forms of GATA1 full length and GATA1 short. (c) Total protein lysates from control infected or RPS19 shRNA infected cells. Molecular weight (MW) markers are shown on the left side of the gel. (d) GATA1 mRNA levels were measured from RPS19 shRNA infected cells (normalized to β-actin; here quantitative RT-PCR results from exons 5–6 are shown with similar results obtained for other exons; n ≥ 3 per group). (e) Polysome profiles from control or RPS19 shRNA treated cells 4 days following infection. The 80S and polysomes are labeled. All graphs are separated on the arbitrary y-axis (derived from relative absorbance at 254 nm) for ease of visualizing the data with the x-axis showing distance along the sucrose gradient. (f) Relative abundance of GATA1 mRNA in polysome fractions as assessed by RT-PCR (normalized to β-actin; *** p < 0.0001). All data are shown as the mean ± the standard deviation (n ≥ 3 per group). (**g**) Relative mRNA abundance in larger polysomes (>4 ribosomes) compared to monosomes. Data are normalized to β-actin. Data are plotted on a log10 scale for ease of viewing the range of relative abundance of various erythroid-important mRNAs. The data are shown as the mean ± the standard deviation (n ≥ 3 per group).
Figure 3. Defects in diverse ribosomal proteins and eukaryotic initiation factors impair GATA1 translation
(a, b, c) Western blots are shown from cells obtained 5 days following infection with shRNAs targeting RPL11, RPL5, and RPS24. Arrowheads indicate the major protein forms of GATA1 full length and GATA1 short. (d) Protein levels after increased concentrations of eIF4E-eIF4G interaction inhibitor 4EGI-1 are used to treat K562 erythroid cells (48 hours of treatment). Arrowheads indicate the major protein forms of GATA1 full length and GATA1 short. (e) Erythropoiesis (as measured by CD235a+ cells) and myeloid cell production (as measured by CD11b+ or CD41a+ cells) following treatment of primary hematopoietic cells with 4EGI-1 (48 hours of treatment). (f) Protein levels after increased concentrations of 4EGI-1 are used to treat CD34-derived primary erythroid cells (48 hours of treatment). Arrowheads indicate the major protein forms of GATA1 full length and GATA1 short.
Figure 4. Global disruption of GATA1 transcriptional activity in DBA patients and rescue of ribosomal protein haploinsufficiency with exogenous GATA1
(a) Mean gene expression values of DBA and control patient samples are plotted on a scatter plot with a linear regression shown in red. The coefficient of determination is shown. (b) Relative GATA1 mRNA levels are plotted from DBA and control samples. (c, d) Enrichment profiles from GSEA comparing the relative expression of genes in DBA patients versus controls and examining the distribution of GATA1 target genes in this data (shown as black bars). (e) Representative FACS plots on transduction of bone marrow mononuclear cells from a DBA patient with GATA1 or HMD (empty) control lentiviruses. (f) The ratio of erythroid (CD235a+) to non-erythroid (CD235a−) -cells is shown after transduction of GATA1 for three independent DBA patients. The data are shown as the mean ± the standard deviation (n = 3). (** p < 0.01; *** p < 0.001). (g) Representative FACS forward scatter histogram plots (measuring cell size) of cultured DBA patient cells transduced with control or GATA1 lentivirus. The mean forward scatter intensity is shown ± the standard deviation (n = 3). (h) Representative cytospin images of cultured DBA patient cells transduced with control or GATA1 lentivirus. Transduced cells were sorted based on GFP expression and stained with May-Grünwald-Giemsa staining. Scale bar = 10 μm. (i) Gene expression analysis by RT-qPCR in DBA patient cells transduced with GATA1 relative to the empty vector control. The data are shown as the mean ± the standard deviation (for 3 independent samples). (Normalized to β-actin; *** p < 0.001)
Comment in
- Reduced translation of GATA1 in Diamond-Blackfan anemia.
Boultwood J, Pellagatti A. Boultwood J, et al. Nat Med. 2014 Jul;20(7):703-4. doi: 10.1038/nm.3630. Nat Med. 2014. PMID: 24999938 No abstract available.
Similar articles
- 5'UTR variants of ribosomal protein S19 transcript determine translational efficiency: implications for Diamond-Blackfan anemia and tissue variability.
Badhai J, Schuster J, Gidlöf O, Dahl N. Badhai J, et al. PLoS One. 2011 Mar 11;6(3):e17672. doi: 10.1371/journal.pone.0017672. PLoS One. 2011. PMID: 21412415 Free PMC article. - Loss of GATA-1 full length as a cause of Diamond-Blackfan anemia phenotype.
Parrella S, Aspesi A, Quarello P, Garelli E, Pavesi E, Carando A, Nardi M, Ellis SR, Ramenghi U, Dianzani I. Parrella S, et al. Pediatr Blood Cancer. 2014 Jul;61(7):1319-21. doi: 10.1002/pbc.24944. Epub 2014 Jan 22. Pediatr Blood Cancer. 2014. PMID: 24453067 Free PMC article. Clinical Trial. - Exome sequencing identifies GATA1 mutations resulting in Diamond-Blackfan anemia.
Sankaran VG, Ghazvinian R, Do R, Thiru P, Vergilio JA, Beggs AH, Sieff CA, Orkin SH, Nathan DG, Lander ES, Gazda HT. Sankaran VG, et al. J Clin Invest. 2012 Jul;122(7):2439-43. doi: 10.1172/JCI63597. Epub 2012 Jun 18. J Clin Invest. 2012. PMID: 22706301 Free PMC article. - Toward RNA Repair of Diamond Blackfan Anemia Hematopoietic Stem Cells.
D'Allard DL, Liu JM. D'Allard DL, et al. Hum Gene Ther. 2016 Oct;27(10):792-801. doi: 10.1089/hum.2016.081. Hum Gene Ther. 2016. PMID: 27550323 Review. - [Connecting isolated congenital asplenia to the ribosome].
Bolze A. Bolze A. Biol Aujourdhui. 2014;208(4):289-98. doi: 10.1051/jbio/2015001. Epub 2015 Apr 3. Biol Aujourdhui. 2014. PMID: 25840456 Review. French.
Cited by
- The Beak of Eukaryotic Ribosomes: Life, Work and Miracles.
Martín-Villanueva S, Galmozzi CV, Ruger-Herreros C, Kressler D, de la Cruz J. Martín-Villanueva S, et al. Biomolecules. 2024 Jul 22;14(7):882. doi: 10.3390/biom14070882. Biomolecules. 2024. PMID: 39062596 Free PMC article. Review. - Decoding ribosome complexity: role of ribosomal proteins in cancer and disease.
Fuentes P, Pelletier J, Gentilella A. Fuentes P, et al. NAR Cancer. 2024 Jul 23;6(3):zcae032. doi: 10.1093/narcan/zcae032. eCollection 2024 Sep. NAR Cancer. 2024. PMID: 39045153 Free PMC article. - GATA1 in Normal and Pathologic Megakaryopoiesis and Platelet Development.
Takasaki K, Chou ST. Takasaki K, et al. Adv Exp Med Biol. 2024;1459:261-287. doi: 10.1007/978-3-031-62731-6_12. Adv Exp Med Biol. 2024. PMID: 39017848 Review. - Alcohol exposure suppresses ribosome biogenesis and causes nucleolar stress in cranial neural crest cells.
Flentke GR, Wilkie TE, Baulch J, Huang Y, Smith SM. Flentke GR, et al. PLoS One. 2024 Jun 28;19(6):e0304557. doi: 10.1371/journal.pone.0304557. eCollection 2024. PLoS One. 2024. PMID: 38941348 Free PMC article. - Towards a Cure for Diamond-Blackfan Anemia: Views on Gene Therapy.
Vale M, Prochazka J, Sedlacek R. Vale M, et al. Cells. 2024 May 27;13(11):920. doi: 10.3390/cells13110920. Cells. 2024. PMID: 38891052 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
- R01 HL107558/HL/NHLBI NIH HHS/United States
- T32 GM007287/GM/NIGMS NIH HHS/United States
- U54 HG003067/HG/NHGRI NIH HHS/United States
- K02 HL111156/HL/NHLBI NIH HHS/United States
- P01 HL032262/HL/NHLBI NIH HHS/United States
- U54 HG003067-09/HG/NHGRI NIH HHS/United States
- P01 HL32262/HL/NHLBI NIH HHS/United States
- R01 HL120791-01/HL/NHLBI NIH HHS/United States
- R01 DK103794/DK/NIDDK NIH HHS/United States
- R21 HL120791/HL/NHLBI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous