Long non-coding RNA HOTAIR, a c-Myc activated driver of malignancy, negatively regulates miRNA-130a in gallbladder cancer - PubMed (original) (raw)
Long non-coding RNA HOTAIR, a c-Myc activated driver of malignancy, negatively regulates miRNA-130a in gallbladder cancer
Ming-Zhe Ma et al. Mol Cancer. 2014.
Abstract
Background: Protein coding genes account for only about 2% of the human genome, whereas the vast majority of transcripts are non-coding RNAs including long non-coding RNAs. A growing volume of literature has proposed that lncRNAs are important players in cancer. HOTAIR was previously shown to be an oncogene and negative prognostic factor in a variety of cancers. However, the factors that contribute to its upregulation and the interaction between HOTAIR and miRNAs are largely unknown.
Methods: A computational screen of HOTAIR promoter was conducted to search for transcription-factor-binding sites. HOTAIR promoter activities were examined by luciferase reporter assay. The function of the c-Myc binding site in the HOTAIR promoter region was tested by a promoter assay with nucleotide substitutions in the putative E-box. The association of c-Myc with the HOTAIR promoter in vivo was confirmed by chromatin immunoprecipitation assay and Electrophoretic mobility shift assay. A search for miRNAs with complementary base paring with HOTAIR was performed utilizing online software program. Gain and loss of function approaches were employed to investigate the expression changes of HOTAIR or miRNA-130a. The expression levels of HOTAIR, c-Myc and miRNA-130a were examined in 65 matched pairs of gallbladder cancer tissues. The effects of HOTAIR and miRNA-130a on gallbladder cancer cell invasion and proliferation was tested using in vitro cell invasion and flow cytometric assays.
Results: We demonstrate that HOTAIR is a direct target of c-Myc through interaction with putative c-Myc target response element (RE) in the upstream region of HOTAIR in gallbladder cancer cells. A positive correlation between c-Myc and HOTAIR mRNA levels was observed in gallbladder cancer tissues. We predicted that HOTAIR harbors a miRNA-130a binding site. Our data showed that this binding site is vital for the regulation of miRNA-130a by HOTAIR. Moreover, a negative correlation between HOTAIR and miRNA-130a was observed in gallbladder cancer tissues. Finally, we demonstrate that the oncogenic activity of HOTAIR is in part through its negative regulation of miRNA-130a.
Conclusion: Together, these results suggest that HOTAIR is a c-Myc-activated driver of malignancy, which acts in part through repression of miRNA-130a.
Figures
Figure 1
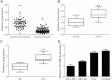
HOTAIR expression in gallbladder cancer and its clinical significance. (A) Difference in expression levels of HOTAIR expression levels between gallbladder cancer tissues and matched non-tumor gallbladder tissues. The expression of HOTAIR was normalized to GADPH. The statistical differences between samples were analyzed with paired samples _t_-test (n = 65, p < 0.0001). (B) Relationship between HOATIR expression and primary tumor growth (p < 0.0001). (C) Relationship between HOATIR expression and lymph node metastasis (p < 0.0001). (D) Expression level of HOTAIR in four gallbladder cancer cell lines. The statistical differences between groups were analyzed using independent samples _t_-test. *p < 0.05; **p < 0.01.
Figure 2
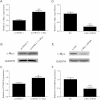
The expression changes of c-Myc or HOTAIR after transfection of c-Myc siRNA or pcDNA3.1-c-Myc in GBC-SD cells. The relative mRNA expression levels were evaluated with real-time qPCR. c-Myc protein levels were determined using Western blot assay. Each experiment was performed in triplicate. (A) pcDNA3.1-c-Myc markedly upregulated the expression of c-Myc at mRNA levels. (B) Representative images of western blot results indicated pcDNA3.1-c-Myc significantly upregulated the expression of c-Myc at protein levels. (C) pcDNA3.1-c-Myc significantly upregulated the expression of HOTAIR at mRNA levels. (D) c-Myc siRNA significantly downregulated the expression of c-Myc at mRNA levels. (E)) Representative images of western blot results indicated c-Myc siRNA significantly downregulated the expression of c-Myc at protein level. (F) c-Myc siRNA significantly down-regulated the expression of HOTAIR at mRNA levels. Error bars represent S.E.M., n = 3. *p < 0.05; **p < 0.01.
Figure 3
c-Myc regulates HOTAIR promoter activity, depending on E-box element. (A) Dual luciferase assay on GBC-SD cells cotransfected with firefly luciferase constructs containing the HOTAIR promoter and pcDNA3.1 or pcDNA3.1-c-Myc. (B) Dual luciferase assay on GBC-SD cells cotransfected with firefly luciferase constructs containing the HOTAIR promoter and c-Myc siRNA or the control siRNA. (C) schematic of the HOTAIR-promoter-luciferase construct is depicted with locations of the E-box element and sequences of point mutation. (D, E) Dual luciferase assay on GBC-SD cells cotransfected with firefly luciferase constructs (mutant at E-box element) and pcDNA3.1-c-Myc (D) or c-Myc siRNA (E). All of the transfection was performed in triplicates. The values are presented as the mean ± S.E.M. of the ratio of firefly luciferase activity to renilla luciferase activity and are representative of at least three independent experiments. Data are shown as the mean ± S.E.M, based on at least three independent experiments. *p < 0.05; **p < 0.01.
Figure 4
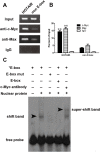
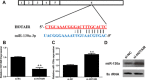
Confirmation of the binding of c-Myc and Max at the promoter region of HOTAIR. (A) c-Myc binding at the promoter region of HOTAIR containing the E-box element and a random region in HOTAIR promoter region (does not contain an E-box, negative control) was assessed by chromatin immunoprecipitation (ChIP). (B) ChIP-derived DNA was amplified by qRT-PCR using specific primers. The levels of qPCR products are expressed as a percentage of input DNA. (C) EMSA showed the interaction of c-Myc with the lncRNA-HOTAIR promoter in vitro. The symbol “*” means the oligonucleotides labled by biotin. Data are shown as the mean ± S.E.M, based on at least three independent experiments. *p < 0.05; **p < 0.01.
Figure 5
Identification of miR-130a as a target of HOTAIR. (A) Alignment of potential HOTAIR base pairing with miR-130a as identified by Starbase v2.0 (
http://starbase.sysu.edu.cn/mirLncRNA.php
). HOATIR (top) consist of 6 exons**,** where the putative binding site is in exon 6. (B) HOTAIR specific siRNA reduced the endogenous HOTAIR mRNA level. (C, D) Upregulation of miR-130a by si-HOTAIR detected by RT-PCR (C) and northern blot (D). GBC-SD cells were transfected with control siRNA or si-HOTAIR, and total RNA was isolated 48 h after transfection. Error bars represent S.E.M., n = 3. *p < 0.05; **p < 0.01.
Figure 6
Identification of miR-130a as a target of HOTAIR. (A) pcDNA3.1-HOTAIR upregulated the HOTAIR mRNA level. (B, C) Downregulation of miR-130a by ectopic expression of HOTAIR detected by RT-PCR (B) and northern blot (C). GBC-SD cells were transfected with vector control or HOTAIR or mutant HOTAIR, and total RNA was isolated 48 h after transfection. (D) The mutant HOTAIR at putative binding site. Error bars represent S.E.M., n = 3. *p < 0.05; **p < 0.01.
Figure 7
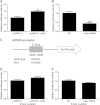
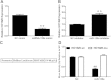
Reciprocal negative regulation of miR-130a and HOTAIR. (A, B) Effect of miR-130a on HOTAIR expression. GBC-SD cells were transfected with vector, miR-130a mimic (A) or miR-130a inhibitor (B), and total RNA was isolated for qRT-PCR 24 h after transfection. (C) Schematic representation of the constructs generated for luciferase assays. (D) HOTAIR WT or its mutant devoid of specific miR-130a-binding sites in which seed matches were mutagenized from ‘TTGTAACGTGA’ to ‘GCGCCUCUUC’ was cloned downstream of Renilla luciferase gene (RLuc) in the vector pRL-TK and transfected into 293 T cells together with specific miRNAs mimics or the negative control mimic (NC). Luciferase assay was performed as described in Materials and Methods. Plotted are results from three independent experiments. Error bars represent S.E.M., n = 3. *p < 0.05; **p < 0.01, n.s., not significant.
Figure 8
The mechanism of the regulation of miR-130a by HOTAIR. (A) Effect of HOTAIR on mature miR-130a, pri-miR-130a and pre-miR-130a. (B) Pull-down of Ago 2 by biotin-labeled HOTAIR or loc285194 RNA probe, as detected by western blot. The loc285194 lane was composed from the same gel with the same contrast. (C) Detection of miR-130a in the same pellet precipitated by the HOTAIR probe, but not in the pellet precipitated by the loc285194 probe. Error bars represent S.E.M., n = 3. *p < 0.05; **p < 0.01, n.s., not significant.
Figure 9
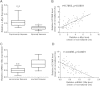
Expression of HOTAIR, c-Myc and miRNA-130a mRNA levels in human gallbladder cancer samples. (A) c-Myc is upregulated in gallbladder cancer tissues compared with paired adjacent normal gallbladder tissues. c-Myc mRNA expression was analyzed by real-time PCR and normalized to GADPH. The statistical differences between samples were analyzed with paired samples _t_-test (n = 65, p < 0.0001). (B) The correlation between HOTAIR and c-Myc expression levels in gallbladder cancer tissues and matched adjacent normal gallbladder tissues (n = 65). Quantitative RT-PCR was performed in triplicate for each sample and assays were repeated once. The relative levels were normalized to GADPH. Each point in the scatter graph represents an individual sample, in which relative c-Myc levels indicate on _x_-axis and HOTAIR levels on _y_-axis. The _x_-axis shows normalized Ct values for c-Myc determined by quantitative RT-PCR. The _y_-axis shows normalized Ct values for HOTAIR determined by quantitative RT-PCR. “Mean of normalized Ct values” is the subtraction of “mean of triple Ct values for c-Myc (_x_-axis) or HOTAIR (_y_-axis)” by “mean of triple Ct values for GADPH”. The correlation coefficient, R = 0.7063, p <0.0001, indicates there is a strongly positive relationship between c-Myc and HOTAIR. (C) miRNA-130a is downregulated in gallbladder cancer tissues compared with paired adjacent normal gallbladder tissues. miRNA-130a mRNA expression was analyzed by real-time PCR and normalized to GADPH. (D) The correlation between HOTAIR and miR-130a expression levels in gallbladder cancer tissues and matched adjacent normal gallbladder tissues (n = 65). The correlation coefficient, R = -0.6398, p <0.0001, indicates there is a strongly negative relationship between miRNA-130a and HOTAIR.
Figure 10
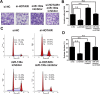
HOTAIR’s oncogenic activity is in part through negative regulation of miRNA-130a. Representative images (A) and the number of migratory cells (B) per high-power field transfected with si-NC, si-HOTAIR, miR-130a inhibitor or si-HOTAIR + miR-130a inhibitor. The migratory ability of GBC-SD cells can be blocked by HOTAIR downregulation. The si-HOTAIR-blocked migratory ability of GBC-SD cells was rescued by miR-130a inhibitor. Quantitative graphical representation of Apoptosis, G1, G2, S cell population (C) and a bar-graphical representation S-Phase Fraction cells in each group (D) transfected with si-NC, si-HOTAIR, miR-130a inhibitor or si-HOTAIR + miR-130a inhibitor. Si-HOTAIR induced a reduction of S-Phase Fraction cells in GBC-SD cells, which can be rescued by miR-130a inhibitor. Error bars represent S.E.M., n = 3. *p < 0.05; **p < 0.01.
Similar articles
- HOTAIR, a long non-coding RNA driver of malignancy whose expression is activated by FOXC1, negatively regulates miRNA-1 in hepatocellular carcinoma.
Su DN, Wu SP, Chen HT, He JH. Su DN, et al. Oncol Lett. 2016 Nov;12(5):4061-4067. doi: 10.3892/ol.2016.5127. Epub 2016 Sep 13. Oncol Lett. 2016. PMID: 27895772 Free PMC article. - Long non-coding RNA CCAT1 promotes gallbladder cancer development via negative modulation of miRNA-218-5p.
Ma MZ, Chu BF, Zhang Y, Weng MZ, Qin YY, Gong W, Quan ZW. Ma MZ, et al. Cell Death Dis. 2015 Jan 8;6(1):e1583. doi: 10.1038/cddis.2014.541. Cell Death Dis. 2015. PMID: 25569100 Free PMC article. - Long noncoding RNA HOTAIR, a hypoxia-inducible factor-1α activated driver of malignancy, enhances hypoxic cancer cell proliferation, migration, and invasion in non-small cell lung cancer.
Zhou C, Ye L, Jiang C, Bai J, Chi Y, Zhang H. Zhou C, et al. Tumour Biol. 2015 Dec;36(12):9179-88. doi: 10.1007/s13277-015-3453-8. Epub 2015 Jun 19. Tumour Biol. 2015. PMID: 26088446 - Intricate crosstalk between MYC and non-coding RNAs regulates hallmarks of cancer.
Swier LJYM, Dzikiewicz-Krawczyk A, Winkle M, van den Berg A, Kluiver J. Swier LJYM, et al. Mol Oncol. 2019 Jan;13(1):26-45. doi: 10.1002/1878-0261.12409. Epub 2018 Dec 5. Mol Oncol. 2019. PMID: 30451365 Free PMC article. Review. - Crosstalk between oncogenic MYC and noncoding RNAs in cancer.
Tu R, Chen Z, Bao Q, Liu H, Qing G. Tu R, et al. Semin Cancer Biol. 2021 Oct;75:62-71. doi: 10.1016/j.semcancer.2020.10.014. Epub 2020 Nov 4. Semin Cancer Biol. 2021. PMID: 33160022 Review.
Cited by
- Overexpression of LncRNA-ROR predicts a poor outcome in gallbladder cancer patients and promotes the tumor cells proliferation, migration, and invasion.
Wang SH, Zhang MD, Wu XC, Weng MZ, Zhou D, Quan ZW. Wang SH, et al. Tumour Biol. 2016 Sep;37(9):12867-12875. doi: 10.1007/s13277-016-5210-z. Epub 2016 Jul 23. Tumour Biol. 2016. PMID: 27449039 - Association between homeobox protein transcript antisense intergenic ribonucleic acid genetic polymorphisms and cholangiocarcinoma.
Lampropoulou DI, Laschos K, Aravantinos G, Georgiou K, Papiris K, Theodoropoulos G, Gazouli M, Filippou D. Lampropoulou DI, et al. World J Clin Cases. 2021 Mar 16;9(8):1785-1792. doi: 10.12998/wjcc.v9.i8.1785. World J Clin Cases. 2021. PMID: 33748227 Free PMC article. - The implications for urological malignancies of non-coding RNAs in the the tumor microenvironment.
Wang S, Qi X, Liu D, Xie D, Jiang B, Wang J, Wang X, Wu G. Wang S, et al. Comput Struct Biotechnol J. 2023 Dec 20;23:491-505. doi: 10.1016/j.csbj.2023.12.016. eCollection 2024 Dec. Comput Struct Biotechnol J. 2023. PMID: 38249783 Free PMC article. Review. - LncRNA HOTAIR epigenetically suppresses miR-122 expression in hepatocellular carcinoma via DNA methylation.
Cheng D, Deng J, Zhang B, He X, Meng Z, Li G, Ye H, Zheng S, Wei L, Deng X, Chen R, Zhou J. Cheng D, et al. EBioMedicine. 2018 Oct;36:159-170. doi: 10.1016/j.ebiom.2018.08.055. Epub 2018 Sep 5. EBioMedicine. 2018. PMID: 30195653 Free PMC article. - Long Non-coding RNA HOTAIR in Central Nervous System Disorders: New Insights in Pathogenesis, Diagnosis, and Therapeutic Potential.
Wang J, Zhao J, Hu P, Gao L, Tian S, He Z. Wang J, et al. Front Mol Neurosci. 2022 Jun 23;15:949095. doi: 10.3389/fnmol.2022.949095. eCollection 2022. Front Mol Neurosci. 2022. PMID: 35813070 Free PMC article. Review.
References
- Jiao Y, Pawlik TM, Anders RA, Selaru FM, Streppel MM, Lucas DJ, Niknafs N, Guthrie VB, Maitra A, Argani P, Offerhaus GJ, Roa JC, Roberts LR, Gores GJ, Popescu I, Alexandrescu ST, Dima S, Fassan M, Simbolo M, Mafficini A, Capelli P, Lawlor RT, Ruzzenente A, Guglielmi A, Tortora G, de Braud F, Scarpa A, Jarnagin W, Klimstra D, Karchin R. et al.Exome sequencing identifies frequent inactivating mutations in BAP1, ARID1A and PBRM1 in intrahepatic cholangiocarcinomas. Nat Genet. 2013;45:1470–1473. doi: 10.1038/ng.2813. - DOI - PMC - PubMed
- Ooi A, Suzuki S, Nakazawa K, Itakura J, Imoto I, Nakamura H, Dobashi Y. Gene amplification of Myc and its coamplification with ERBB2 and EGFR in gallbladder adenocarcinoma. Anticancer Res. 2009;29:19–26. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical