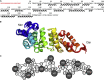Elongated structure of the outer-membrane activator of peptidoglycan synthesis LpoA: implications for PBP1A stimulation - PubMed (original) (raw)
Elongated structure of the outer-membrane activator of peptidoglycan synthesis LpoA: implications for PBP1A stimulation
Nicolas L Jean et al. Structure. 2014.
Abstract
The bacterial cell envelope contains the stress-bearing peptidoglycan layer, which is enlarged during cell growth and division by membrane-anchored synthases guided by cytoskeletal elements. In Escherichia coli, the major peptidoglycan synthase PBP1A requires stimulation by the outer-membrane-anchored lipoprotein LpoA. Whereas the C-terminal domain of LpoA interacts with PBP1A to stimulate its peptide crosslinking activity, little is known about the role of the N-terminal domain. Herein we report its NMR structure, which adopts an all-α-helical fold comprising a series of helix-turn-helix tetratricopeptide-repeat (TPR)-like motifs. NMR spectroscopy of full-length LpoA revealed two extended flexible regions in the C-terminal domain and limited, if any, flexibility between the N- and C-terminal domains. Analytical ultracentrifugation and small-angle X-ray scattering results are consistent with LpoA adopting an elongated shape, with dimensions sufficient to span from the outer membrane through the periplasm to interact with the peptidoglycan synthase PBP1A.
Copyright © 2014 Elsevier Ltd. All rights reserved.
Figures
Graphical abstract
Figure 1
LpoAN Has a TPR Domain-like Structure (A) Amino acid sequence of the LpoAN construct used for structure determination and secondary structure elements (α helices). The residues of the oligohistidine tag are shown in red. (B) Cartoon representation of the 20 lowest-energy structures of LpoAN, as determined by NMR spectroscopy, emphasizing the spatial arrangement of the 12 α-helices, which are numbered from the N to C terminus. (C) LpoAN is stabilized by numerous interhelical hydrophobic contacts. The ten α helices with more than four residues are shown as circles and labeled according to the description in (B). Interhelical hydrophobic contacts are represented with dashed lines, and short plain lines connect each of the residues to its corresponding helix. See also Figure S1.
Figure 2
Comparison of LpoAC from E. coli and H. influenzae (A) Superimposition of the X-ray structure of H. influenzae LpoAC (green, PDB code
3CKM
) and the structure of E. coli LpoAC predicted by PHYRE (red). Regions 1 (residues N285–P351) and 2 (residues S493–N531) are present in LpoAC from E. coli, but not from H. influenzae. Flexible residues for which backbone resonances have been assigned by NMR spectroscopy are sketched as blue sticks. (B) Disorder in E. coli LpoAC predicted by IUPred. The two main regions absent from the H. influenzae LpoA sequence (in gray) are predicted as being mostly unstructured by IUPred (score > 0.5). (C) Sequence alignment of H. influenzae and E. coli LpoAC, where the two E. coli inserts are highlighted in gray. NMR-assigned residues are shown in blue. (D) The linker between LpoAN and LpoAC (in gray and white hashes) starts at K249 and ends at K258. The criteria used to define this linker included the structuring of the N-terminal domain as quantified by the {1H}15N-NOE measured in LpoAN (black) and the definition of the first secondary-structure element in the C-terminal domain (red) of LpoA as modeled by PHYRE. See also Figure S2.
Figure 3
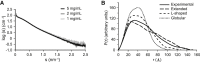
Full-Length LpoA Has an Extended Structure (A) SAXS curves of LpoA at concentrations of 1, 2, and 5 mg/ml. (B) Experimental distance distribution function, P(r), calculated from SAXS data collected on a 5 mg/ml 15N-LpoA sample (black) and theoretical P(r) function calculated for molecular models with three different, arbitrarily chosen orientations of the N- and C-terminal domains (....., globular model; ----, L-shaped model; – – –, extended model; see also Figure S3C). The theoretical Rg values extracted from these curves are 3.11 nm for the globular model, 3.58 nm for the L-shaped model, and 4.44 nm for the extended model. The experimental Rg value, 4.22 ± 0.01 nm, fits best to the extended model.
Figure 4
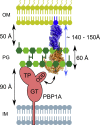
Schematic Representation of PBP1A Activation by LpoA The N-terminal domain of LpoA (blue) anchors to the outer membrane, whereas the C-terminal domain (orange) interacts with the outer-membrane PBP1A docking domain (Typas et al., 2010). LpoA has an estimated total width of ∼30 Å and length of ∼145 Å. These dimensions should enable the protein to reach PBP1A through the periplasm and cross the ∼60-Å-thick PG layer, which has ∼40- to 60-Å-wide pores (Demchick and Koch, 1996). TP, transpeptidase domain; GT, glycosyltransferase domain; IM, inner membrane; OM, outer membrane.
Similar articles
- Activities and regulation of peptidoglycan synthases.
Egan AJ, Biboy J, van't Veer I, Breukink E, Vollmer W. Egan AJ, et al. Philos Trans R Soc Lond B Biol Sci. 2015 Oct 5;370(1679):20150031. doi: 10.1098/rstb.2015.0031. Philos Trans R Soc Lond B Biol Sci. 2015. PMID: 26370943 Free PMC article. Review. - Crystal structures of the amino-terminal domain of LpoA from Escherichia coli and Haemophilus influenzae.
Kelley A, Vijayalakshmi J, Saper MA. Kelley A, et al. Acta Crystallogr F Struct Biol Commun. 2019 May 1;75(Pt 5):368-376. doi: 10.1107/S2053230X19004011. Epub 2019 Apr 26. Acta Crystallogr F Struct Biol Commun. 2019. PMID: 31045566 Free PMC article. - The LpoA activator is required to stimulate the peptidoglycan polymerase activity of its cognate cell wall synthase PBP1a.
Sardis MF, Bohrhunter JL, Greene NG, Bernhardt TG. Sardis MF, et al. Proc Natl Acad Sci U S A. 2021 Aug 31;118(35):e2108894118. doi: 10.1073/pnas.2108894118. Proc Natl Acad Sci U S A. 2021. PMID: 34429361 Free PMC article. - Structural analyses of the Haemophilus influenzae peptidoglycan synthase activator LpoA suggest multiple conformations in solution.
Sathiyamoorthy K, Vijayalakshmi J, Tirupati B, Fan L, Saper MA. Sathiyamoorthy K, et al. J Biol Chem. 2017 Oct 27;292(43):17626-17642. doi: 10.1074/jbc.M117.804997. Epub 2017 Sep 8. J Biol Chem. 2017. PMID: 28887305 Free PMC article. - Lipoproteins: Structure, Function, Biosynthesis.
Braun V, Hantke K. Braun V, et al. Subcell Biochem. 2019;92:39-77. doi: 10.1007/978-3-030-18768-2_3. Subcell Biochem. 2019. PMID: 31214984 Review.
Cited by
- Lytic transglycosylase MltG cleaves in nascent peptidoglycan and produces short glycan strands.
Sassine J, Pazos M, Breukink E, Vollmer W. Sassine J, et al. Cell Surf. 2021 May 1;7:100053. doi: 10.1016/j.tcsw.2021.100053. eCollection 2021 Dec. Cell Surf. 2021. PMID: 34036206 Free PMC article. - Interaction of lipopolysaccharides at intermolecular sites of the periplasmic Lpt transport assembly.
Laguri C, Sperandeo P, Pounot K, Ayala I, Silipo A, Bougault CM, Molinaro A, Polissi A, Simorre JP. Laguri C, et al. Sci Rep. 2017 Aug 29;7(1):9715. doi: 10.1038/s41598-017-10136-0. Sci Rep. 2017. PMID: 28852068 Free PMC article. - Activities and regulation of peptidoglycan synthases.
Egan AJ, Biboy J, van't Veer I, Breukink E, Vollmer W. Egan AJ, et al. Philos Trans R Soc Lond B Biol Sci. 2015 Oct 5;370(1679):20150031. doi: 10.1098/rstb.2015.0031. Philos Trans R Soc Lond B Biol Sci. 2015. PMID: 26370943 Free PMC article. Review. - Tandem-repeat protein domains across the tree of life.
Jernigan KK, Bordenstein SR. Jernigan KK, et al. PeerJ. 2015 Jan 13;3:e732. doi: 10.7717/peerj.732. eCollection 2015. PeerJ. 2015. PMID: 25653910 Free PMC article. - Escherichia coli has robust regulatory mechanisms against elevated peptidoglycan cleavage by lytic transglycosylases.
Liang Y, Zhao Y, Kwan JMC, Wang Y, Qiao Y. Liang Y, et al. J Biol Chem. 2023 Apr;299(4):104615. doi: 10.1016/j.jbc.2023.104615. Epub 2023 Mar 16. J Biol Chem. 2023. PMID: 36931392 Free PMC article.
References
- Banzhaf M., van den Berg van Saparoea B., Terrak M., Fraipont C., Egan A., Philippe J., Zapun A., Breukink E., Nguyen-Distèche M., den Blaauwen T., Vollmer W. Cooperativity of peptidoglycan synthases active in bacterial cell elongation. Mol. Microbiol. 2012;85:179–194. - PubMed
- Bertsche U., Breukink E., Kast T., Vollmer W. In vitro murein peptidoglycan synthesis by dimers of the bifunctional transglycosylase-transpeptidase PBP1B from Escherichia coli. J. Biol. Chem. 2005;280:38096–38101. - PubMed
- Born P., Breukink E., Vollmer W. In vitro synthesis of cross-linked murein and its attachment to sacculi by PBP1A from Escherichia coli. J. Biol. Chem. 2006;281:26985–26993. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases