RNF2 is recruited by WASH to ubiquitinate AMBRA1 leading to downregulation of autophagy - PubMed (original) (raw)
RNF2 is recruited by WASH to ubiquitinate AMBRA1 leading to downregulation of autophagy
Pengyan Xia et al. Cell Res. 2014 Aug.
Abstract
WASH (Wiskott-Aldrich syndrome protein (WASP) and SCAR homolog) was identified to function in endosomal sorting via Arp2/3 activation. We previously demonstrated that WASH is a new interactor of BECN1 and present in the BECN1-PIK3C3 complex with AMBRA1. The AMBRA1-DDB1-CUL4A complex is an E3 ligase for K63-linked ubiquitination of BECN1, which is required for starvation-induced autophagy. WASH suppresses autophagy by inhibition of BECN1 ubiquitination. However, how AMBRA1 is regulated during autophagy remains elusive. Here, we found that RNF2 associates with AMBRA1 to act as an E3 ligase to ubiquitinate AMBRA1 via K48 linkage. RNF2 mediates ubiquitination of AMBRA1 at lysine 45. Notably, RNF2 deficiency enhances autophagy induction. Upon autophagy induction, RNF2 potentiates AMBRA1 degradation with the help of WASH. WASH deficiency impairs the association of RNF2 with AMBRA1 to impede AMBRA1 degradation. Our findings reveal another novel layer of regulation of autophagy through WASH recruitment of RNF2 for AMBRA1 degradation leading to downregulation of autophagy.
Figures
Figure 1
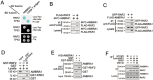
RNF2 interacts with AMBRA1. (A) RNF2 interacts with AMBRA1 in yeast two-hybrid assays. Yeast strain AH109 was transfected with BD (Gal4 DNA-binding domain)-fused RNF2 and AD (Gal4 activating domain)-fused AMBRA1 and plated on the indicated medium for 3 d. TP53 with large T antigen was introduced as a positive control. (B, C) RNF2 interacts with AMBRA1 in mammalian cells. FLAG-tagged RNF2 and MYC-tagged AMBRA1 were co-transfected into MEF cells for 24 h, followed by IP with antibody against FLAG (B). GFP-tagged RNF2 and FLAG-tagged AMBRA1 were co-transfected into MEF cells for 24 h, followed by IP with antibody against FLAG (C). (D) GST-RNF2 precipitates AMBRA1 from MEF cell lysates. GST-RNF2 or GST was bound to GST beads and incubated with MEF cell lysates, followed by immunoblotting with the indicated antibodies. (E) RNF2 directly interacts with AMBRA1. Recombinant GST-tagged RNF2 were incubated with HIS-AMBRA1, followed by GST pull-down assays. (F) Endogenous RNF2 interacts with AMBRA1 upon starvation. MEFs were treated with culture medium (CM) or EBSS for 2 h, followed by IP with antibody against RNF2. 10 μM MG132 was added to cell culture before EBSS treatment. Experiments were repeated three times with similar results.
Figure 2
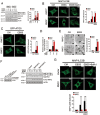
RNF2 suppresses autophagy. (A) RNF2 deficiency leads to enhanced autophagic activity. WT control (RNF2+/+) and RNF2 KO (RNF2−/−) MEFs were starved in EBSS for 4 h with or without BafA1. Lysates were analyzed by immunoblotting with the indicated antibodies (left panel) and the ratio of MAP1LC3B-II/ACTB was calculated (right panel). (B) MAP1LC3B dots were increased in RNF2 KO cells. RNF2+/+ and RNF2−/− MEF cells were starved in EBSS for 4 h with or without BafA1. Endogenous MAP1LC3B was stained with antibody against MAP1LC3B and visualized by confocal microscopy (left panel). The number of MAP1LC3B dots/puncta was counted from at least 100 cells for each condition (right panel). Scale bar, 10 μm. (C) ATG5 dots are increased in RNF2−/− MEFs. RNF2+/+ and RNF2−/− MEFs stably expressing GFP-ATG5 were stimulated with EBSS for 4 h. GFP-ATG5 dots were visualized and counted as B. (D) RNF2 deficiency increases degradation of long-lived proteins in MEFs under starvation conditions. 3-MA, a type III phosphatidylinositol 3-kinases (PI-3K) inhibitor, was added to block the activity of autophagy. (E) RNF2-deficient MEFs exhibit enhanced autophagy. RNF2+/+ and RNF2−/− MEFs were treated with culture medium (CM) or EBSS for 4 h, followed by examination with transmission electron microscopy. Black arrowhead indicates autophagosome. The number of autophagosomes was counted (right panel). Scale bar, 1 μm. (F) RNF2 restoration inhibits autophagy. RNF2−/− MEFs stably expressing WT or H69Y-RNF2 mutant (left panel) were treated with EBSS for 4 h. Lysates were analyzed by immunoblotting with the indicated antibodies (right panel). (G) RNF2 overexpression decreases MAP1LC3B puncta. MEF cells were transfected with control vector or RNF2 followed by culture in EBSS or CM for 4 h with or without BafA1. MAP1LC3B dots were analyzed as B. Scale bar, 10 μm. Data are representative of three independent experiments and shown as means ± SD. **P< 0.01; ***P< 0.001.
Figure 3
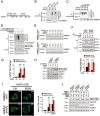
RNF2 promotes AMBRA1 degradation through a proteasome degradation pathway during autophagy. (A) RNF2 deficiency prevents degradation of AMBRA1. RNF2+/+ and RNF2−/− MEFs treated with EBSS for the indicated times were harvested for immunoblotting (left panel). Ratios of AMBRA1/ACTB were calculated (right panel). (B) The half-life of AMBRA1. RNF2+/+ (upper panel) and RNF2−/− (lower panel) MEFs treated with or without EBSS for the indicated times in the presence of 20 μg/ml cycloheximide (CHX) were harvested for immunoblotting with the indicated antibodies. (C) RNF2 deficiency does not change the mRNA level of AMBRA1. RNF2+/+ and RNF2−/− MEFs were harvested, and mRNA levels for the indicated genes were analyzed by RT-PCR. mRNA levels of AMBRA1 were normalized to those of ACTB (right panel). (D) RNF2 restoration in RNF2−/− MEFs rescues the degradation of AMBRA1 during autophagy. RNF2−/− MEFs were infected with lentiviruses encoding WT- or H69Y-RNF2 and treated with EBSS, followed by immunoblotting (left panel). Ratios of AMBRA1/ACTB were calculated (right panel). (E, F) AMBRA1 is degraded through a proteasome degradation pathway. MEFs were treated with the indicated reagents (20 μg/ml CHX, 20 μM CQ, 10 μM MG132) for 6 h in CM (E) or EBSS (F), and harvested for immunoblotting. Data are representative of at least three separate experiments and shown as means ± SD.
Figure 4
RNF2 is an E3 ligase for K48-linked ubiquitination of AMBRA1. (A, B) RNF2 promotes degradation of AMBRA1. FLAG-tagged AMBRA1 and increasing amounts of MYC-tagged RNF2 were co-transfected into MEF cells for 24 h. The protein levels were examined by immunoblotting with the indicated antibodies (A). The mRNA levels were examined by RT-PCR with primers specific for exogenous plasmids (B, left panel**). mRNA levels of AMBRA1 were normalized to those of GFP (B, right panel)**. (C) RNF2 overexpression enhances K48-linked ubiquitination of AMBRA1. RNF2−/− MEFs transfected with the indicated RNF2 plasmids and ubiquitin mutants were starved in EBSS for 2 h in the presence of 10 μM MG132, followed by IP with antibody against AMBRA1. The immunoprecipitates were dissociated with 1% SDS and reimmunoprecipitated with anti-AMBRA1. Immunoprecipitates and lysates were immunoblotted with the indicated antibodies. K63-polyubiquitinated BECN1 was immunoprecipitated with antibody against BECN1 from starved cells stably expressing HA-Ub as a positive control for antibody against K63-linked polyubiquitin chains. The mRNA level of AMBRA1 was examined by RT-PCR. (D) RNF2 ubiquitinates AMBRA1 via K48 linkage by in vitro ubiquitination reconstitution assays. Recombinant RNF2 was incubated with purified FLAG-AMBRA1 in the presence of recombinant E1 (UBA1) and E2 (UBE2D3) and ubiquitin mutants at 30 °C for 1 h, followed by IP with antibody against FLAG and immunoblotting with the indicated antibodies. (E) RNF2 deficiency blocks the ubiquitination of AMBRA1 during autophagy. RNF2+/+ and RNF2−/− MEFs transfected with various ubiquitin mutants were starved in EBSS for 2 h in the presence of 10 μM MG132, followed by IP with antibody against AMBRA1. Data represent at least three separate experiments.
Figure 5
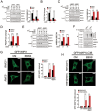
K45R-AMBRA1 mutant maintains AMBRA1 stability and promotes PIK3C3 activity. (A) Scheme for lysines on AMBRA1. (B, C) K45 is the RNF2-mediated ubiquitination site on AMBRA1. Various AMBRA1 truncations (B) or mutants (C) were co-transfected with RNF2 with or without HA-tagged ubiquitin (HA-Ub) into MEFs for 24 h. Cells were harvested after pretreatment with 10 μM MG132 for 6 h and EBSS for 2 h, followed by IP with antibody against AMBRA1. The immunoprecipitates were dissociated with 1% SDS and reimmunoprecipitated with anti-FLAG. Immunoprecipitates were immunoblotted with the indicated antibodies. (D) K45R-AMBRA1 enhances autophagy. AMBRA1-silenced MEFs were stably rescued with WT-AMBRA1 or K45R-AMBRA1 and starved in EBSS for the indicated times, followed by IP with antibody against FLAG after pretreated with 10 μM MG132 for 6 h. Immunoprecipitates were immunoblotted with the indicated antibodies. (E) The half-life of AMBRA1. AMBRA1-silenced MEFs were transfected with WT-AMBRA1 (upper panel) or K45R-AMBRA1 (lower panel) and selected for stable expression with 2 μg/ml puromycin. Cells were then treated with or without EBSS for the indicated times in the presence of 20 μg/ml CHX before harvest for immunoblotting with the indicated antibodies. (F) The association of BECN1 with PIK3C3 is enhanced in K45R-AMBRA1-rescued cells. AMBRA1-silenced MEFs were rescued with WT-AMBRA1 or K45R-AMBRA1 for 24 h, followed by IP with anti-BECN1 antibody. Immunoprecipitates were immunoblotted with the indicated antibodies (upper panel). The ratios of PIK3C3 versus BECN1 were calculated and shown in the lower panel. (G) K45R-AMBRA1 augments PIK3C3 kinase activity. AMBRA1-silenced MEFs were rescued with WT-AMBRA1 or K45R-AMBRA1, followed by starvation in EBSS for the indicated times. Cells were lysed for immunoblotting with anti-ATG14 antibody for autophagy-specific PIK3C3. Immunoprecipitates were split into two equal parts, one for a loading control and the other for in vitro kinase assay. Radioactive PI(3)P was resolved by thin layer chromatography (TLC), and normalized against the BECN1-associated PIK3C3. (H, I) K45R-AMBRA1 promotes autophagic activity. AMBRA1-silenced MEFs were rescued with WT-AMBRA1 or K45R-AMBRA1, followed by starvation in EBSS for 2 h with or without 20 nM BafA1. Cells were lysed for immunoblotting with the indicated antibodies (H). Endogenous MAP1LC3B were stained with antibody against MAP1LC3B and MAP1LC3B dots were counted (I). (J) RNF2 deficiency does not affect the K45R-AMBRA1-elevated autophagy. AMBRA1-silenced MEFs were stably rescued with WT-AMBRA1 or K45R-AMBRA1, followed by starvation in EBSS for 2 h with or without 20 nM BafA1. Cells were lysed for immunoblotting with the indicated antibodies. Data are representative of at least three separate experiments and shown as means ± SD. Scale bar, 10 μm. **P< 0.01; ***P< 0.001.
Figure 6
RNF2 inhibits autophagy through promoting AMBRA1 degradation. (A) RNF2 suppresses the association between BECN1 and PIK3C3. MEFs transfected with vector or RNF2 were starved in EBSS for the indicated times, followed by IP with anti-BECN1 antibody. Ratios of PIK3C3 versus BECN1 were calculated and shown in the right panel. (B) RNF2 inhibits the PIK3C3 kinase activity. RNF2-overexpressing MEFs were starved in EBSS for the indicated times, followed by IP with anti-ATG14 antibody for autophagy-specific PIK3C3. Immunoprecipitates were split into two equal parts and analyzed as Figure 5G. (C) RNF2 deficiency enhances the association between BECN1 and PIK3C3. RNF2+/+ and RNF2−/− MEFs were starved in EBSS for the indicated times, followed by IP with anti-BECN1 antibody and assayed as A. (D) RNF2 deficiency promotes the PIK3C3 kinase activity. RNF2+/+ and RNF2−/− MEFs were starved in EBSS for the indicated times, and harvested for IP with anti-ATG14 antibody for autophagy-specific PIK3C3. Immunoprecipitates were analyzed as B. (E) RNF2 deficiency increases the amount of AMBRA1 that is associated with BECN1. RNF2+/+ and RNF2−/− MEFs were incubated with EBSS for the indicated times and harvested for IP with anti-BECN1 antibody (left panel). Ratios of AMBRA1/BECN1 were analyzed and shown in the right panel. (F) RNF2 deficiency increases K63-linked ubiquitination of BECN1 during autophagy. RNF2+/+ and RNF2−/− MEFs were starved in EBSS for the indicated times, and harvested for IP with antibody against BECN1. Immunoprecipitates and cell lysates were immunoblotted with the indicated antibodies. (G) RNF2 deficiency elevates the kinase activity of PIK3C3. RNF2+/+ and RNF2−/− MEFs stably expressing GFP-WIPI1 were starved in EBSS for 2 h with or without BafA1 treatment. GFP-WIPI1 dots were counted and shown in the right panel. (H) H69Y-RNF2 mutation abolishes the inhibition of autophagy. MEFs stably expressing GFP-MAP1LC3B were transfected with WT-RNF2 or H69Y-RNF2 mutant, followed by starvation in EBSS for 2 h with or without BafA1 treatment. GFP-MAP1LC3B dots were counted and are shown in the lower panel. Data are representative of at least three separate experiments and shown as means ± SD. Scale bar, 10 μm. **P< 0.01; ***P< 0.001.
Figure 7
WASH mediates RNF2 recruitment for AMBRA1 degradation. (A) WASH interacts with RNF2 in yeast two-hybrid assays. Yeast strain AH109 was transfected with BD-fused WASH and AD-fused RNF2 and plated on the indicated medium for 3 d. TP53 with large T antigen or a known WASH interactor Bloc1s2 (data not shown) was introduced as positive controls. (B) WASH directly interacts with RNF2. Recombinant WASH (rWASH) and GST-RNF2 were subjected to GST pull-down assays. (C) WASH enhances the interaction between RNF2 and AMBRA1. GST-RNF2 or GST was incubated with recombinant AMBRA1 with or without WASH, followed by GST pull-down assays. (D) RNF2 associates with the components of the BECN1-PIK3C3 complex. Cell lysates from MEFs treated with or without EBSS for 2 h were immunoprecipitated with anti-RNF2 antibody. (E, F) WASH promotes degradation of AMBRA1. MEFs transfected with MYC-AMBRA1 and increasing amounts of FLAG-WASH were harvested 24 h post-transfection for immunoblotting with the indicated antibodies (E). mRNAs were extracted and subjected to RT-PCR analysis with primers specific for exogenous plasmids (F). GFP served as a transfection control. (G) K45R-AMBRA1 mutation abolishes WASH-augmented degradation of AMBRA1. HA-tagged WASH was transfected into AMBRA1-depleted MEFs rescued with WT- or K45R-AMBRA1 for 24 h. Cells were starved in EBSS for 4 h, and harvested for immunoblotting with the indicated antibodies. (H) WASH deficiency prevents degradation of AMBRA1. WASH+/+ or WASH−/− MEFs treated with 20 μg/ml CHX plus EBSS for the indicated times were harvested for immunoblotting (left panel). Blot band intensity ratios of AMBRA1/ACTB were calculated and shown as means ± SD (right panel). (I) AMBRA1 associates with RNF2 dependently of WASH. WASH+/+ or WASH−/− MEFs were treated with or without EBSS for 2 h, and cell lysates were immunoprecipitated with anti-RNF2 antibody. (J) WASH deficiency impairs polyubiquitination of AMBRA1. WASH+/+ or WASH−/− MEFs were starved in EBSS for the indicated times, followed by IP with antibody against AMBRA1 after pretreatment with 10 μM MG132 for 6 h. Immunoprecipitates were immunoblotted with the indicated antibodies. (K) WASH promotes polyubiquitination of AMBRA1 in vitro. Recombinant RNF2 was incubated with purified FLAG-AMBRA1, E1 (UBA1), E2 (UBE2D3) and ubiquitin mutants with or without recombinant WASH at 30 °C for 1 h, followed by IP with antibody against FLAG and immunoblotting with the indicated antibodies. Experiments were repeated three times with similar results.
Comment in
- Autophagy regulation: RNF2 targets AMBRA1.
Morel E, Dupont N, Codogno P. Morel E, et al. Cell Res. 2014 Sep;24(9):1029-30. doi: 10.1038/cr.2014.105. Epub 2014 Aug 8. Cell Res. 2014. PMID: 25104734 Free PMC article.
Similar articles
- Autophagy regulation: RNF2 targets AMBRA1.
Morel E, Dupont N, Codogno P. Morel E, et al. Cell Res. 2014 Sep;24(9):1029-30. doi: 10.1038/cr.2014.105. Epub 2014 Aug 8. Cell Res. 2014. PMID: 25104734 Free PMC article. - WASH inhibits autophagy through suppression of Beclin 1 ubiquitination.
Xia P, Wang S, Du Y, Zhao Z, Shi L, Sun L, Huang G, Ye B, Li C, Dai Z, Hou N, Cheng X, Sun Q, Li L, Yang X, Fan Z. Xia P, et al. EMBO J. 2013 Oct 16;32(20):2685-96. doi: 10.1038/emboj.2013.189. Epub 2013 Aug 23. EMBO J. 2013. PMID: 23974797 Free PMC article. - Autophagy induction in atrophic muscle cells requires ULK1 activation by TRIM32 through unanchored K63-linked polyubiquitin chains.
Di Rienzo M, Antonioli M, Fusco C, Liu Y, Mari M, Orhon I, Refolo G, Germani F, Corazzari M, Romagnoli A, Ciccosanti F, Mandriani B, Pellico MT, De La Torre R, Ding H, Dentice M, Neri M, Ferlini A, Reggiori F, Kulesz-Martin M, Piacentini M, Merla G, Fimia GM. Di Rienzo M, et al. Sci Adv. 2019 May 8;5(5):eaau8857. doi: 10.1126/sciadv.aau8857. eCollection 2019 May. Sci Adv. 2019. PMID: 31123703 Free PMC article. - AMBRA1, a Novel BH3-Like Protein: New Insights Into the AMBRA1-BCL2-Family Proteins Relationship.
Di Rita A, Strappazzon F. Di Rita A, et al. Int Rev Cell Mol Biol. 2017;330:85-113. doi: 10.1016/bs.ircmb.2016.09.002. Epub 2016 Nov 15. Int Rev Cell Mol Biol. 2017. PMID: 28215535 Review. - Impaired autophagy and APP processing in Alzheimer's disease: The potential role of Beclin 1 interactome.
Salminen A, Kaarniranta K, Kauppinen A, Ojala J, Haapasalo A, Soininen H, Hiltunen M. Salminen A, et al. Prog Neurobiol. 2013 Jul-Aug;106-107:33-54. doi: 10.1016/j.pneurobio.2013.06.002. Epub 2013 Jul 1. Prog Neurobiol. 2013. PMID: 23827971 Review.
Cited by
- Systematic Discovery of Human Gene Function and Principles of Modular Organization through Phylogenetic Profiling.
Dey G, Jaimovich A, Collins SR, Seki A, Meyer T. Dey G, et al. Cell Rep. 2015 Feb 17;10(6):993-1006. doi: 10.1016/j.celrep.2015.01.025. Epub 2015 Feb 12. Cell Rep. 2015. PMID: 25683721 Free PMC article. - The Roles of Ubiquitin in Mediating Autophagy.
Yin Z, Popelka H, Lei Y, Yang Y, Klionsky DJ. Yin Z, et al. Cells. 2020 Sep 2;9(9):2025. doi: 10.3390/cells9092025. Cells. 2020. PMID: 32887506 Free PMC article. Review. - Linc01194 acts as an oncogene in colorectal carcinoma and is associated with poor survival outcome.
Wang X, Liu Z, Tong H, Peng H, Xian Z, Li L, Hu B, Xie S. Wang X, et al. Cancer Manag Res. 2019 Mar 22;11:2349-2362. doi: 10.2147/CMAR.S189189. eCollection 2019. Cancer Manag Res. 2019. PMID: 30962722 Free PMC article. - Neurodevelopmental Disorders: Functional Role of Ambra1 in Autism and Schizophrenia.
La Barbera L, Vedele F, Nobili A, D'Amelio M, Krashia P. La Barbera L, et al. Mol Neurobiol. 2019 Oct;56(10):6716-6724. doi: 10.1007/s12035-019-1557-7. Epub 2019 Mar 26. Mol Neurobiol. 2019. PMID: 30915711 Review. - E3 ubiquitin ligase RNF2 protects polymerase ι from destabilization.
Fedorowicz M, Halas A, Macias M, Sledziewska-Gojska E, Woodgate R, McIntyre J. Fedorowicz M, et al. Biochim Biophys Acta Mol Cell Res. 2024 Jun;1871(5):119743. doi: 10.1016/j.bbamcr.2024.119743. Epub 2024 May 3. Biochim Biophys Acta Mol Cell Res. 2024. PMID: 38705361
References
- Mizushima N, Komatsu M. Autophagy: renovation of cells and tissues. Cell. 2011;147:728–741. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases