Spore formation in Bacillus subtilis - PubMed (original) (raw)
Review
Spore formation in Bacillus subtilis
Irene S Tan et al. Environ Microbiol Rep. 2014 Jun.
Abstract
Although prokaryotes ordinarily undergo binary fission to produce two identical daughter cells, some are able to undergo alternative developmental pathways that produce daughter cells of distinct cell morphology and fate. One such example is a developmental programme called sporulation in the bacterium Bacillus subtilis, which occurs under conditions of environmental stress. Sporulation has long been used as a model system to help elucidate basic processes of developmental biology including transcription regulation, intercellular signalling, membrane remodelling, protein localization and cell fate determination. This review highlights some of the recent work that has been done to further understand prokaryotic cell differentiation during sporulation and its potential applications.
Published 2013. This article is a U.S. Government work and is in the public domain in the USA.
Figures
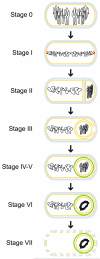
Figure 1
Schematic representation of morphological changes that occur during sporulation in Bacillus subtilis. Distinct stages of sporulation are denoted with a Roman numeral, according to the numbering scheme proposed by Ryter (Ryter, 1965). Peptidoglycan is depicted in gray, membranes are depicted in yellow, DNA is depicted in black, the position of the origin of replication of the chromosomes is shown as a red dot at stage 0 and I, and the spore coat is depicted in green. At stage 0, chromosomes are replicated, but no obvious morphological landmarks of sporulation are yet present. Stage I is defined by chromosome condensation and the anchoring of the origins of replication to the extreme poles of the cell. In stage II, the polar septum is elaborated, followed by engulfment of the forespore in stage III. Stage IV and V represent cortex and coat assembly, respectively. Stage VI refers to “spore maturation”; a particularly obvious morphological feature elaborated at this stage is the tightly condensed, toroidal structure of the forespore chromosome. In stage VII, the mother cell lyses, releasing the mature, largely dormant spore into the environment.
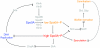
Figure 2
Genetic circuitry that governs the entry into sporulation. Arrows indicate activation; repression is denoted by a bar. Developmental events are depicted in the color corresponding to Spo0A levels or phosphorylation states that govern that event. Thus, unphosphorylated Spo0A corresponds to active DNA replication; low levels of phosphorylated Spo0A (SpoA~P) leads to biofilm formation and cannabilistic behavior; and high levels of phosphorylated Spo0A drives the entry into sporulation. Proteins other than Spo0A that participate in each activation or repression step are depicted in gray.
Similar articles
- A model for asymmetric septum formation during sporulation in Bacillus subtilis.
Errington J. Errington J. Mol Microbiol. 1991 Apr;5(4):785-9. doi: 10.1111/j.1365-2958.1991.tb00750.x. Mol Microbiol. 1991. PMID: 1906965 Review. - Visualizing Bacillus subtilis During Vegetative Growth and Spore Formation.
Wang X, Montero Llopis P. Wang X, et al. Methods Mol Biol. 2016;1431:275-87. doi: 10.1007/978-1-4939-3631-1_19. Methods Mol Biol. 2016. PMID: 27283315 - Regulation of endospore formation in Bacillus subtilis.
Errington J. Errington J. Nat Rev Microbiol. 2003 Nov;1(2):117-26. doi: 10.1038/nrmicro750. Nat Rev Microbiol. 2003. PMID: 15035041 Review. - Differentiation of Vegetative Cells into Spores: a Kinetic Model Applied to Bacillus subtilis.
Gauvry E, Mathot AG, Couvert O, Leguérinel I, Jules M, Coroller L. Gauvry E, et al. Appl Environ Microbiol. 2019 May 2;85(10):e00322-19. doi: 10.1128/AEM.00322-19. Print 2019 May 15. Appl Environ Microbiol. 2019. PMID: 30902849 Free PMC article. - Sporulation in Bacillus subtilis. Effect of medium on the form of chromosome replication and on initiation to sporulation in Bacillus subtilis.
Mandelstam J, Sterlini JM, Kay D. Mandelstam J, et al. Biochem J. 1971 Nov;125(2):635-41. doi: 10.1042/bj1250635. Biochem J. 1971. PMID: 5004200 Free PMC article.
Cited by
- A versatile nano display platform from bacterial spore coat proteins.
Wu IL, Narayan K, Castaing JP, Tian F, Subramaniam S, Ramamurthi KS. Wu IL, et al. Nat Commun. 2015 Apr 9;6:6777. doi: 10.1038/ncomms7777. Nat Commun. 2015. PMID: 25854653 Free PMC article. - Effects of drying strategies on sporulation and titer of microbial ecological agents with Bacillus subtilis.
Li C, Zhao K, Ma L, Zhao J, Zhao ZM. Li C, et al. Front Nutr. 2022 Sep 27;9:1025248. doi: 10.3389/fnut.2022.1025248. eCollection 2022. Front Nutr. 2022. PMID: 36238457 Free PMC article. - Reformulation of an extant ATPase active site to mimic ancestral GTPase activity reveals a nucleotide base requirement for function.
Updegrove TB, Harke J, Anantharaman V, Yang J, Gopalan N, Wu D, Piszczek G, Stevenson DM, Amador-Noguez D, Wang JD, Aravind L, Ramamurthi KS. Updegrove TB, et al. Elife. 2021 Mar 11;10:e65845. doi: 10.7554/eLife.65845. Elife. 2021. PMID: 33704064 Free PMC article. - Mechanisms and Applications of Bacterial Sporulation and Germination in the Intestine.
Koopman N, Remijas L, Seppen J, Setlow P, Brul S. Koopman N, et al. Int J Mol Sci. 2022 Mar 21;23(6):3405. doi: 10.3390/ijms23063405. Int J Mol Sci. 2022. PMID: 35328823 Free PMC article. Review. - Differential requirements for conserved peptidoglycan remodeling enzymes during Clostridioides difficile spore formation.
Ribis JW, Fimlaid KA, Shen A. Ribis JW, et al. Mol Microbiol. 2018 Nov;110(3):370-389. doi: 10.1111/mmi.14090. Mol Microbiol. 2018. PMID: 30066347 Free PMC article.
References
- Ben-Yehuda S, Fujita M, Liu XS, Gorbatyuk B, Skoko D, Yan J, et al. Defining a centromere-like element in Bacillus subtilis by Identifying the binding sites for the chromosome-anchoring protein RacA. Mol Cell. 2005;17:773–782. - PubMed
- Ben-Yehuda S, Rudner DZ, Losick R. RacA, a bacterial protein that anchors chromosomes to the cell poles. Science. 2003;299:532–536. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources