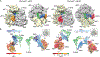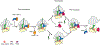Cryoelectron microscopic structures of eukaryotic translation termination complexes containing eRF1-eRF3 or eRF1-ABCE1 - PubMed (original) (raw)
Cryoelectron microscopic structures of eukaryotic translation termination complexes containing eRF1-eRF3 or eRF1-ABCE1
Anne Preis et al. Cell Rep. 2014.
Abstract
Termination and ribosome recycling are essential processes in translation. In eukaryotes, a stop codon in the ribosomal A site is decoded by a ternary complex consisting of release factors eRF1 and guanosine triphosphate (GTP)-bound eRF3. After GTP hydrolysis, eRF3 dissociates, and ABCE1 can bind to eRF1-loaded ribosomes to stimulate peptide release and ribosomal subunit dissociation. Here, we present cryoelectron microscopic (cryo-EM) structures of a pretermination complex containing eRF1-eRF3 and a termination/prerecycling complex containing eRF1-ABCE1. eRF1 undergoes drastic conformational changes: its central domain harboring the catalytically important GGQ loop is either packed against eRF3 or swung toward the peptidyl transferase center when bound to ABCE1. Additionally, in complex with eRF3, the N-terminal domain of eRF1 positions the conserved NIKS motif proximal to the stop codon, supporting its suggested role in decoding, yet it appears to be delocalized in the presence of ABCE1. These results suggest that stop codon decoding and peptide release can be uncoupled during termination.
Copyright © 2014 The Authors. Published by Elsevier Inc. All rights reserved.
Figures
Figure 1.. Cryo-EM Structures of Pretermination and Termination/Prerecycling Complexes
(A) Side and top views of the 80S ribosome pretermination complex with eRF1 and eRF3 (left) and termination/prerecycling complex with eRF1-ABCE1 (right). Density attributed to eRF1 occupies the A site. In the termination/prerecycling complex, the position of the flexible NTD of eRF1 is outlined with a black line. (B) Molecular models for peptidyl tRNA, eRF1, eRF3, and ABCE1 on the ribosome. The NIKS motif (pink spheres) of eRF1 is positioned in close proximity to the stop codon (orange). The central domain of eRF1 containing the GGQ loop (magenta spheres) is packed against eRF3. In complex with ABCE1, the central domain of eRF1 is swung toward the PTC.
Figure 2.. eRF1-Ribosome Interactions and Positioning of the NTD of eRF1 in the Pretermination Complex
(A) eRF1 forms multiple contacts with the ribosome (left) that are mostly identical to those of Pelota in complex with Hbs1 (right) (Becker et al., 2011), apart from a contact at h8-h14 of the 18S rRNA. The minidomain of the CTD of eRF1 contacts ES8 and S31 near the beak of the SSU. (B) The NTD reaches deep into the decoding center and establishes multiple contacts with 18S rRNA and S12 (left). The NIKS motif is close to the stop codon in the A site (orange). (C) For decoding of the stop codon, bacterial RF1 and RF2 (Korostelev et al., 2008; Laurberg et al., 2008) rely on domain II that is unrelated to eRF1 NTD. Interacting amino acids are marked in pink.
Figure 3.. eRF1 Interactions and Positioning of Its Central Domain in the Termination/Prerecycling Complex
(A) The central domain of eRF1 undergoes a conformational change that positions the GGQ loop near the CCA end of the P site-tRNA (left). The CTD moves away from the SSU and forms contacts with the stalk base of the LSU and the SRL. These conformational changes are very similar to those of Pelota in complex with ABCE1 (middle). Unrelated domain III of bacterial RF1 possesses a different architecture but coordinates the highly conserved GGQ loop in an identical position (right). (B) Cross-section and close-up view of the central domain of eRF1 with the GGQ loop close to the peptidyl tRNA (left and middle). Position and conformation of the GGQ loop are highly similar to that of bacterial RF1 (Laurberg et al., 2008).
Figure 4.. Scheme of Eukaryotic Translation Termination and Ribosome Recycling
For termination, the stop codon in the A site is recognized by the eRF1-eRF3-GTP ternary complex. eRF3 dissociates after GTP hydrolysis and allows the central domain of eRF1 to swing to the PTC. Proper positioning of the GGQ motif in the central domain of eRF1 may already allow peptide release, resulting in a termination complex with the deacyl-tRNA in the P state or P/E hybrid state. Alternatively, the active conformation of eRF1 in the pretermination complex is stabilized after binding of ABCE1. This stimulates peptide release while the NTD of eRF1 is delocalized, thus decoupling decoding from peptide release. Independent of the termination mechanism, ABCE1 together with eRF1 functions in concert to dissociate the ribosome into small and large subunits.
Similar articles
- Cryo-EM structure of the mammalian eukaryotic release factor eRF1-eRF3-associated termination complex.
Taylor D, Unbehaun A, Li W, Das S, Lei J, Liao HY, Grassucci RA, Pestova TV, Frank J. Taylor D, et al. Proc Natl Acad Sci U S A. 2012 Nov 6;109(45):18413-8. doi: 10.1073/pnas.1216730109. Epub 2012 Oct 22. Proc Natl Acad Sci U S A. 2012. PMID: 23091004 Free PMC article. - Translation termination depends on the sequential ribosomal entry of eRF1 and eRF3.
Beißel C, Neumann B, Uhse S, Hampe I, Karki P, Krebber H. Beißel C, et al. Nucleic Acids Res. 2019 May 21;47(9):4798-4813. doi: 10.1093/nar/gkz177. Nucleic Acids Res. 2019. PMID: 30873535 Free PMC article. - Structural view on recycling of archaeal and eukaryotic ribosomes after canonical termination and ribosome rescue.
Franckenberg S, Becker T, Beckmann R. Franckenberg S, et al. Curr Opin Struct Biol. 2012 Dec;22(6):786-96. doi: 10.1016/j.sbi.2012.08.002. Epub 2012 Sep 29. Curr Opin Struct Biol. 2012. PMID: 23031510 Review. - Translation Termination and Ribosome Recycling in Eukaryotes.
Hellen CUT. Hellen CUT. Cold Spring Harb Perspect Biol. 2018 Oct 1;10(10):a032656. doi: 10.1101/cshperspect.a032656. Cold Spring Harb Perspect Biol. 2018. PMID: 29735640 Free PMC article. Review.
Cited by
- The Structural Dynamics of Translation.
Korostelev AA. Korostelev AA. Annu Rev Biochem. 2022 Jun 21;91:245-267. doi: 10.1146/annurev-biochem-071921-122857. Epub 2022 Mar 14. Annu Rev Biochem. 2022. PMID: 35287473 Free PMC article. Review. - Ribosome recycling is coordinated by processive events in two asymmetric ATP sites of ABCE1.
Nürenberg-Goloub E, Heinemann H, Gerovac M, Tampé R. Nürenberg-Goloub E, et al. Life Sci Alliance. 2018 Jun 14;1(3):e201800095. doi: 10.26508/lsa.201800095. Life Sci Alliance. 2018. PMID: 30198020 Free PMC article. - Polyadenylate-binding protein-interacting proteins PAIP1 and PAIP2 affect translation termination.
Ivanov A, Shuvalova E, Egorova T, Shuvalov A, Sokolova E, Bizyaev N, Shatsky I, Terenin I, Alkalaeva E. Ivanov A, et al. J Biol Chem. 2019 May 24;294(21):8630-8639. doi: 10.1074/jbc.RA118.006856. Epub 2019 Apr 16. J Biol Chem. 2019. PMID: 30992367 Free PMC article. - A structural inventory of native ribosomal ABCE1-43S pre-initiation complexes.
Kratzat H, Mackens-Kiani T, Ameismeier M, Potocnjak M, Cheng J, Dacheux E, Namane A, Berninghausen O, Herzog F, Fromont-Racine M, Becker T, Beckmann R. Kratzat H, et al. EMBO J. 2021 Jan 4;40(1):e105179. doi: 10.15252/embj.2020105179. Epub 2020 Dec 8. EMBO J. 2021. PMID: 33289941 Free PMC article. - Suppression of Nonsense Mutations by New Emerging Technologies.
Morais P, Adachi H, Yu YT. Morais P, et al. Int J Mol Sci. 2020 Jun 20;21(12):4394. doi: 10.3390/ijms21124394. Int J Mol Sci. 2020. PMID: 32575694 Free PMC article. Review.
References
- Alkalaeva EZ, Pisarev AV, Frolova LY, Kisselev LL, and Pestova TV. (2006). In vitro reconstitution of eukaryotic translation reveals cooperativity between release factors eRF1 and eRF3. Cell 125, 1125–1136. - PubMed
- Berninghausen O, and Beckmann R. (2011). Structure of the no-go mRNA decay complex Dom34-Hbs1 bound to a stalled 80S ribosome. Nat. Struct. Mol. Biol 18, 715–720. - PubMed
- Bhushan S, Meyer H, Starosta AL, Becker T, Mielke T, Berninghausen O, Sattler M, Wilson DN, and Beckmann R. (2010). Structural basis for translational stalling by human cytomegalovirus and fungal arginine attenuator peptide. Mol. Cell 40, 138–146. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases