Evolutionary genomics of fast evolving tunicates - PubMed (original) (raw)
Review
Evolutionary genomics of fast evolving tunicates
Luisa Berná et al. Genome Biol Evol. 2014.
Abstract
Tunicates have been extensively studied because of their crucial phylogenetic location (the closest living relatives of vertebrates) and particular developmental plan. Recent genome efforts have disclosed that tunicates are also remarkable in their genome organization and molecular evolutionary patterns. Here, we review these latter aspects, comparing the similarities and specificities of two model species of the group: Oikopleura dioica and Ciona intestinalis. These species exhibit great genome plasticity and Oikopleura in particular has undergone a process of extreme genome reduction and compaction that can be explained in part by gene loss, but is mostly due to other mechanisms such as shortening of intergenic distances and introns, and scarcity of mobile elements. In Ciona, genome reorganization was less severe being more similar to the other chordates in several aspects. Rates and patterns of molecular evolution are also peculiar in tunicates, being Ciona about 50% faster than vertebrates and Oikopleura three times faster. In fact, the latter species is considered as the fastest evolving metazoan recorded so far. Two processes of increase in evolutionary rates have taken place in tunicates. One of them is more extreme, and basically restricted to genes encoding regulatory proteins (transcription regulators, chromatin remodeling proteins, and metabolic regulators), and the other one is less pronounced but affects the whole genome. Very likely adaptive evolution has played a very significant role in the first, whereas the functional and/or evolutionary causes of the second are less clear and the evidence is not conclusive. The evidences supporting the incidence of increased mutation and less efficient negative selection are presented and discussed.
Keywords: Ciona; Oikopleura dioica; genome plasticity; positive selection.
© The Author(s) 2014. Published by Oxford University Press on behalf of the Society for Molecular Biology and Evolution.
Figures
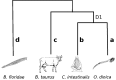
Fig. 1.—
HOX gene cluster evolution. Schematic representation of the HOX cluster evolution in representative Eumetazoan groups. Hox genes are schematized as follows: anterior Hox genes (yellow), paralogy group 3 Hox genes (orange), central Hox genes (green), and posterior Hox genes (blue). Horizontal lines connecting genes indicate linkage. The lancelet cluster is considered as the canonical one, since it is complete and did not undergo rearrangements (Monteiro and Ferrier 2006). Tunicates lack some of the central Hox genes and the cluster is broken. Specifically, Ciona intestinalis lost four Hox genes (Hox 7, 8, 9, and 11) and the Hox cluster was separated in five syntenic segments, whereas Oikopleura dioica lost five Hox genes (3, 5, 6, 7, and 8) and the cluster is completely disintegrated, namely none of the genes kept synteny (Seo et al. 2004). Complex rearrangements of Hox gene order occurred in sea urchin genome (Cameron et al. 2006). However, other echinodermes present temporal and spatial collinearity for several Hox genes (Mooi and David 2008). Although some insects present a collinear Hox cluster, this is partially fragmented in Drosophila melanogaster (genes also referred to as: Lab, pb, z2, zen, bcd, Dfd, Scr, ftz, and Antp corresponding to the first cluster and Ubx, abd-A, and Abd-B to the second one) (Hughes and Kaufman 2002).
Fig. 2.—
Different mechanisms of genome reduction: (A) Gene loss, (B) operon organization, (C) reduction of intergenic regions, and (D) intronic regions shrink. The arrow pointing upward next to the species name indicates that the mechanism is more pronounce in that species.
Fig. 3.—
Schematic representation depicting the phylogenetic relationships of the species used to estimate the relative rate of molecular evolution on the basis of branch lengths. D1 stands for the length of the branch connecting the common ancestors of tunicates with that of tunicates and vertebrates (i.e., Olfactores); a, b, and c the lengths of the respective branches.
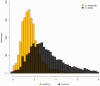
Fig. 4.—
Distribution of (b + D1)/c and (a + D1)/c ratios for Ciona intestinalis (yellow) and Oikopleura dioica (dark gray). The values a, b, D1, and c correspond to branch lengths as schematized in figure 3. More specifically, a and b correspond to the branch lengths between O. dioica and Ciona to their common ancestor, respectively, and c the branch length between Bos taurus and its common ancestor with tunicates.
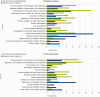
Fig. 5.—
Gene ontology (GO) enrichment analysis of highly accelerated genes. Representative GO terms that exhibit statistically significant differences are shown in the graphic (Fisher’s exact test, FDR; P < 0.05). Distribution of GO terms for the 300 most accelerated ((a + D1)/c >5) genes from Oikopleura dioica (A) and ((b + D1)/c >2.5) Ciona intestinalis (B) and the respective reference sets (remainder of the genome). The values a, b, D1, and c correspond to the branch lengths for O. dioica, Ciona, and Bos taurus to the respective common ancestors, as depicted in figure 3. Modified from Berná et al. (2012).
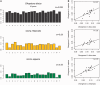
Fig. 6.—
(A) Proportion of divergent positions refers to the fraction of nonconserved positions for each amino acid between the extant species and the ancestor of reference (for cases of Oikopleura dioica and Ciona intestinalis the ancestor would be that of tunicates). The pattern in human is shown for reference. CV stands for the coefficient of variation (i.e., variance over mean) in substitution rates. (B) Scatter plot of amino acid divergence patterns in the group of slow evolving genes from Oikopleura versus the pattern observed in Ciona and human. The lowest panel is a scatter plot of amino acid divergence patterns between Ciona and human included to illustrate the degree of similarity that “normal” divergence patterns exhibit.
Similar articles
- Evolutionary changes in the notochord genetic toolkit: a comparative analysis of notochord genes in the ascidian Ciona and the larvacean Oikopleura.
Kugler JE, Kerner P, Bouquet JM, Jiang D, Di Gregorio A. Kugler JE, et al. BMC Evol Biol. 2011 Jan 20;11:21. doi: 10.1186/1471-2148-11-21. BMC Evol Biol. 2011. PMID: 21251251 Free PMC article. - Peculiar patterns of amino acid substitution and conservation in the fast evolving tunicate Oikopleura dioica.
Berná L, D'Onofrio G, Alvarez-Valin F. Berná L, et al. Mol Phylogenet Evol. 2012 Feb;62(2):708-17. doi: 10.1016/j.ympev.2011.11.013. Epub 2011 Nov 23. Mol Phylogenet Evol. 2012. PMID: 22120064 - Tunicates and not cephalochordates are the closest living relatives of vertebrates.
Delsuc F, Brinkmann H, Chourrout D, Philippe H. Delsuc F, et al. Nature. 2006 Feb 23;439(7079):965-8. doi: 10.1038/nature04336. Nature. 2006. PMID: 16495997 - THALIACEANS, THE NEGLECTED PELAGIC RELATIVES OF ASCIDIANS: A DEVELOPMENTAL AND EVOLUTIONARY ENIGMA.
Piette J, Lemaire P. Piette J, et al. Q Rev Biol. 2015 Jun;90(2):117-45. doi: 10.1086/681440. Q Rev Biol. 2015. PMID: 26285352 Review.
Cited by
- Genome-Wide Identification, Comparison, and Expression Analysis of Transcription Factors in Ascidian Styela clava.
Zhang J, Wei J, Yu H, Dong B. Zhang J, et al. Int J Mol Sci. 2021 Apr 21;22(9):4317. doi: 10.3390/ijms22094317. Int J Mol Sci. 2021. PMID: 33919240 Free PMC article. - Hidden diversity in Antarctica: Molecular and morphological evidence of two different species within one of the most conspicuous ascidian species.
Ruiz MB, Taverna A, Servetto N, Sahade R, Held C. Ruiz MB, et al. Ecol Evol. 2020 Jul 15;10(15):8127-8143. doi: 10.1002/ece3.6504. eCollection 2020 Aug. Ecol Evol. 2020. PMID: 32788966 Free PMC article. - Distinguishing Evolutionary Conservation from Derivedness.
Leong JCK, Uesaka M, Irie N. Leong JCK, et al. Life (Basel). 2022 Mar 17;12(3):440. doi: 10.3390/life12030440. Life (Basel). 2022. PMID: 35330191 Free PMC article. Review. - TIAMMAt: Leveraging Biodiversity to Revise Protein Domain Models, Evidence from Innate Immunity.
Tassia MG, David KT, Townsend JP, Halanych KM. Tassia MG, et al. Mol Biol Evol. 2021 Dec 9;38(12):5806-5818. doi: 10.1093/molbev/msab258. Mol Biol Evol. 2021. PMID: 34459919 Free PMC article. - Identification and population genetic comparison of three ascidian species based on mtDNA sequences.
Bhattachan P, Qiao R, Dong B. Bhattachan P, et al. Ecol Evol. 2020 Mar 10;10(8):3758-3768. doi: 10.1002/ece3.6171. eCollection 2020 Apr. Ecol Evol. 2020. PMID: 32313634 Free PMC article.
References
- Berná L, D’Onofrio G, Alvarez-Valin F. Peculiar patterns of amino acid substitution and conservation in the fast evolving tunicate Oikopleura dioica. Mol Phylogenet Evol. 2012;62:708–717. - PubMed
- Bourlat SJ, et al. Deuterostome phylogeny reveals monophyletic chordates and the new phylum Xenoturbellida. Nature. 2006;444:85–88. - PubMed
- Byrd J, Lambert CC. Mechanism of the block to hybridization and selfing between the sympatric ascidians Ciona intestinalis and Ciona savignyi. Mol Reprod Dev. 2000;56:541. - PubMed
- Cameron RA, et al. Unusual gene order and organization of the sea urchin hox cluster. Exp Zool B Mol Dev Evol. 2006;306:45–58. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources