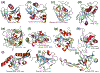The natural history of ADP-ribosyltransferases and the ADP-ribosylation system - PubMed (original) (raw)
Review
The natural history of ADP-ribosyltransferases and the ADP-ribosylation system
L Aravind et al. Curr Top Microbiol Immunol. 2015.
Abstract
Catalysis of NAD(+)-dependent ADP-ribosylation of proteins, nucleic acids, or small molecules has evolved in at least three structurally unrelated superfamilies of enzymes, namely ADP-ribosyltransferase (ART), the Sirtuins, and probably TM1506. Of these, the ART superfamily is the most diverse in terms of structure, active site residues, and targets that they modify. The primary diversification of the ART superfamily occurred in the context of diverse bacterial conflict systems, wherein ARTs play both offensive and defensive roles. These include toxin-antitoxin systems, virus-host interactions, intraspecific antagonism (polymorphic toxins), symbiont/parasite effectors/toxins, resistance to antibiotics, and repair of RNAs cleaved in conflicts. ARTs evolving in these systems have been repeatedly acquired by lateral transfer throughout eukaryotic evolution, starting from the PARP family, which was acquired prior to the last eukaryotic common ancestor. They were incorporated into eukaryotic regulatory/epigenetic control systems (e.g., PARP family and NEURL4), and also used as defensive (e.g., pierisin and CARP-1 families) or immunity-related proteins (e.g., Gig2-like ARTs). The ADP-ribosylation system also includes other domains, such as the Macro, ADP-ribosyl glycohydrolase, NADAR, and ADP-ribosyl cyclase, which appear to have initially diversified in bacterial conflict-related systems. Unlike ARTs, sirtuins appear to have a much smaller presence in conflict-related systems.
Figures
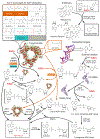
Fig. 1
Summary of known ADP-ribosylation targets and pathways. Most substrates are tagged by ADP-ribose by different members of the ADP-ribosyltransferase fold (ARTs and PARPs). Cysteine and arginine residues (orange) may also serve as substrates for reactions catalyzed by members of the sirtuin family and aspartate (turquoise) is possibly a substrate of the TM1506 family. The enzyme responsible for phosphoserine (gray) ADP-ribosylation remains unknown. At the bottom of the figure, some key reactions for processing ADP-ribose derivatives are represented. ART ADP-ribosyltransferase, PARP poly(ADP-ribosyl) polymerase, CDPase 2′-cyclic phosphate hydrolase, KptA/Tpt1 RNA 2′-phosphotransferase (ART fold), Pierisin lepidopteran cytotoxin (ART fold), ARH ADP-ribosylglycohydrolase, PARG poly(ADP-ribosyl)-glycohydrolase, Macro ADP-ribose associated Macro domain, Nudix “nucleotide diphosphate linked to X” hydrolase domain. Sirtuins might deacylate proteins with several distinct acyl groups (show as R-), such as acetyl, malonyl, crotonyl, succinyl or palmitoyl groups
Fig. 2
Reconstructed evolutionary history for the different members of ART superfamily. Horizontal lines are colored according to their observed phyletic distributions (key: bottom right). Dashes indicate uncertainty in terms of the origins of a family, while the gray ellipse indicates that the RolB/6b clade likely underwent rapid divergence from either the H-Y-[EDQ] or R-S-E clade. The H-H-h and H-Y-[EDQ] include the ARTD proteins, while the R-S-E clade includes the ARTC proteins in the previously presented nomenclature for ART domains (Hottiger et al. 2010). For each family the known or inferred functional role in biological conflicts or processes is indicated (right column). Representatives of each family are shown in Figs. 4 and 5. In the bottom panel shows the idealized topology of the ART fold with markup indicating specific structural features associated with certain ART families and positions of active site residues
Fig. 3
Cartoon structures of diverse members of the ART superfamily. Conserved strands and helices of the core fold are shown in aquamarine and red, respectively. Additional strands that pack with the sheets and inserts are shown in wheat color. Conserved strands of the core are labeled, as also active site residues and ligands. a KptA (PDB: 1wfx). b CC0527 (PDB: 2o0q). c Rifampin ART (PDB: 2hw2). d BC2332 (PDB: 2aua). e PARP (PDB: 1uk1). f Pseudomonas exotoxin A (PDB: 3B8H). g _β_-NAD+ glycohydrolyase (PDB: 4KT6A). h 6b (RolB) (PDB: 3aq3). i Pertussis toxin (PDB: 1prt). j Ecto-ART (PDB: log1). k AvrPphF (PDB: 1s21)
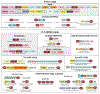
Fig. 4
Domain architectures and gene neighborhoods of ARTs of the H-H-h and H-Y-[EDQ] clades. Gene neighborhoods and domain architectures are labeled with the gene name, Genbank index (gi) number and the species name, which is shown in brackets. For domain architectures, the label corresponds to the ART containing gene. Gene neighborhoods are shown as box-arrows with the arrowhead pointing to the gene in the 3′ direction of the coding strand. These are further shaded gray. The architectures are grouped based on the type of ART. Standard domain abbreviations are used to label genes and domains
Fig. 5
Domain architectures and gene neighborhoods of ARTs of the R-S-E clade. The labeling scheme and grouping of proteins and gene neighborhoods are as described in Fig. 4. Standard domain abbreviations are used to label genes and domains. Additionally, X and Y are uncharacterized domains
Similar articles
- Identification of novel components of NAD-utilizing metabolic pathways and prediction of their biochemical functions.
de Souza RF, Aravind L. de Souza RF, et al. Mol Biosyst. 2012 Jun;8(6):1661-77. doi: 10.1039/c2mb05487f. Epub 2012 Mar 7. Mol Biosyst. 2012. PMID: 22399070 - Rapid evolution of PARP genes suggests a broad role for ADP-ribosylation in host-virus conflicts.
Daugherty MD, Young JM, Kerns JA, Malik HS. Daugherty MD, et al. PLoS Genet. 2014 May 29;10(5):e1004403. doi: 10.1371/journal.pgen.1004403. eCollection 2014. PLoS Genet. 2014. PMID: 24875882 Free PMC article. - Players in ADP-ribosylation: Readers and Erasers.
Verheugd P, Bütepage M, Eckei L, Lüscher B. Verheugd P, et al. Curr Protein Pept Sci. 2016;17(7):654-667. doi: 10.2174/1389203717666160419144846. Curr Protein Pept Sci. 2016. PMID: 27090904 Review. - Mono(ADP-ribosyl)transferases and related enzymes in animal tissues. Emerging gene families.
Koch-Nolte F, Haag F. Koch-Nolte F, et al. Adv Exp Med Biol. 1997;419:1-13. doi: 10.1007/978-1-4419-8632-0_1. Adv Exp Med Biol. 1997. PMID: 9193631 Review. - ADP-ribosylation of arginine.
Laing S, Unger M, Koch-Nolte F, Haag F. Laing S, et al. Amino Acids. 2011 Jul;41(2):257-69. doi: 10.1007/s00726-010-0676-2. Epub 2010 Jul 21. Amino Acids. 2011. PMID: 20652610 Free PMC article. Review.
Cited by
- (ADP-ribosyl)hydrolases: structure, function, and biology.
Rack JGM, Palazzo L, Ahel I. Rack JGM, et al. Genes Dev. 2020 Mar 1;34(5-6):263-284. doi: 10.1101/gad.334631.119. Epub 2020 Feb 6. Genes Dev. 2020. PMID: 32029451 Free PMC article. Review. - Photorhabdus antibacterial Rhs polymorphic toxin inhibits translation through ADP-ribosylation of 23S ribosomal RNA.
Jurėnas D, Payelleville A, Roghanian M, Turnbull KJ, Givaudan A, Brillard J, Hauryliuk V, Cascales E. Jurėnas D, et al. Nucleic Acids Res. 2021 Aug 20;49(14):8384-8395. doi: 10.1093/nar/gkab608. Nucleic Acids Res. 2021. PMID: 34255843 Free PMC article. - Polyvalent Proteins, a Pervasive Theme in the Intergenomic Biological Conflicts of Bacteriophages and Conjugative Elements.
Iyer LM, Burroughs AM, Anand S, de Souza RF, Aravind L. Iyer LM, et al. J Bacteriol. 2017 Jul 11;199(15):e00245-17. doi: 10.1128/JB.00245-17. Print 2017 Aug 1. J Bacteriol. 2017. PMID: 28559295 Free PMC article. - What the Wild Things Do: Mechanisms of Plant Host Manipulation by Bacterial Type III-Secreted Effector Proteins.
Schreiber KJ, Chau-Ly IJ, Lewis JD. Schreiber KJ, et al. Microorganisms. 2021 May 11;9(5):1029. doi: 10.3390/microorganisms9051029. Microorganisms. 2021. PMID: 34064647 Free PMC article. Review. - The chemistry of the vitamin B3 metabolome.
Makarov MV, Trammell SAJ, Migaud ME. Makarov MV, et al. Biochem Soc Trans. 2019 Feb 28;47(1):131-147. doi: 10.1042/BST20180420. Epub 2018 Dec 17. Biochem Soc Trans. 2019. PMID: 30559273 Free PMC article. Review.
References
- Adriouch S, Ohlrogge W, Haag F, Koch-Nolte F, Seman M (2001) Rapid induction of naive T cell apoptosis by ecto-nicotinamide adenine dinucleotide: requirement for mono(ADP-ribosyl)transferase 2 and a downstream effector. J Immunol 167(1):196–203 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials