Recovery of a medieval Brucella melitensis genome using shotgun metagenomics - PubMed (original) (raw)
Recovery of a medieval Brucella melitensis genome using shotgun metagenomics
Gemma L Kay et al. mBio. 2014.
Abstract
Shotgun metagenomics provides a powerful assumption-free approach to the recovery of pathogen genomes from contemporary and historical material. We sequenced the metagenome of a calcified nodule from the skeleton of a 14th-century middle-aged male excavated from the medieval Sardinian settlement of Geridu. We obtained 6.5-fold coverage of a Brucella melitensis genome. Sequence reads from this genome showed signatures typical of ancient or aged DNA. Despite the relatively low coverage, we were able to use information from single-nucleotide polymorphisms to place the medieval pathogen genome within a clade of B. melitensis strains that included the well-studied Ether strain and two other recent Italian isolates. We confirmed this placement using information from deletions and IS711 insertions. We conclude that metagenomics stands ready to document past and present infections, shedding light on the emergence, evolution, and spread of microbial pathogens. Importance: Infectious diseases have shaped human populations and societies throughout history. The recovery of pathogen DNA sequences from human remains provides an opportunity to identify and characterize the causes of individual and epidemic infections. By sequencing DNA extracted from medieval human remains through shotgun metagenomics, without target-specific capture or amplification, we have obtained a draft genome sequence of an ~700-year-old Brucella melitensis strain. Using a variety of bioinformatic approaches, we have shown that this historical strain is most closely related to recent strains isolated from Italy, confirming the continuity of this zoonotic infection, and even a specific lineage, in the Mediterranean region over the centuries.
Copyright © 2014 Kay et al.
Figures
FIG 1
(A) Calcific nodules excavated from the pelvic girdle of skeleton 2658. (B) Size distribution of reads from sample 2568. (i) All reads from the initial MiSeq run (~2 m). (ii) Reads from the second Miseq run (>20 m) which aligned with the B. melitensis 16 M genome. (iii) Reads from the dedicated run which mapped to the human genome (hg19). (C) Coverage plot of reads from sample 2568 mapped against the two chromosomes from the B. melitensis 16 M genome. The plot shows the average coverage and standard deviation for each 5,000-bp region in the genome. (D) Evidence of sequence damage associated with aged DNA showing the frequency of C-to-T and G-to-A transitions at the 5′ and 3′ ends of DNA fragments, respectively.
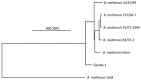
FIG 2
Phylogenetic tree showing the position of the medieval Geridu-1 strain within the Ether clade. Only those SNPs which correlated with sufficient coverage in the Geridu-1 alignment were included in the construction of the tree.
Comment in
- Ancient-pathogen genomics: coming of age?
Whatmore AM. Whatmore AM. mBio. 2014 Sep 2;5(5):e01676-14. doi: 10.1128/mBio.01676-14. mBio. 2014. PMID: 25182326 Free PMC article.
Similar articles
- Full genome SNP-based phylogenetic analysis reveals the origin and global spread of Brucella melitensis.
Tan KK, Tan YC, Chang LY, Lee KW, Nore SS, Yee WY, Mat Isa MN, Jafar FL, Hoh CC, AbuBakar S. Tan KK, et al. BMC Genomics. 2015 Feb 18;16(1):93. doi: 10.1186/s12864-015-1294-x. BMC Genomics. 2015. PMID: 25888205 Free PMC article. - Ancient-pathogen genomics: coming of age?
Whatmore AM. Whatmore AM. mBio. 2014 Sep 2;5(5):e01676-14. doi: 10.1128/mBio.01676-14. mBio. 2014. PMID: 25182326 Free PMC article. - Examining pathogen DNA recovery across the remains of a 14th century Italian friar (Blessed Sante) infected with Brucella melitensis.
Hider J, Duggan AT, Klunk J, Eaton K, Long GS, Karpinski E, Giuffra V, Ventura L, Fornaciari A, Fornaciari G, Golding GB, Prowse TL, Poinar HN. Hider J, et al. Int J Paleopathol. 2022 Dec;39:20-34. doi: 10.1016/j.ijpp.2022.08.002. Epub 2022 Sep 26. Int J Paleopathol. 2022. PMID: 36174312 - The genome of Brucella melitensis.
DelVecchio VG, Kapatral V, Elzer P, Patra G, Mujer CV. DelVecchio VG, et al. Vet Microbiol. 2002 Dec 20;90(1-4):587-92. doi: 10.1016/s0378-1135(02)00238-9. Vet Microbiol. 2002. PMID: 12414174 Review. - Infectious disease in the Pleistocene: Old friends or old foes?
Houldcroft CJ, Underdown S. Houldcroft CJ, et al. Am J Biol Anthropol. 2023 Dec;182(4):513-531. doi: 10.1002/ajpa.24737. Epub 2023 Mar 30. Am J Biol Anthropol. 2023. PMID: 38006200 Review.
Cited by
- The Recovery, Interpretation and Use of Ancient Pathogen Genomes.
Duchêne S, Ho SYW, Carmichael AG, Holmes EC, Poinar H. Duchêne S, et al. Curr Biol. 2020 Oct 5;30(19):R1215-R1231. doi: 10.1016/j.cub.2020.08.081. Curr Biol. 2020. PMID: 33022266 Free PMC article. Review. - Culture-independent detection and characterisation of Mycobacterium tuberculosis and M. africanum in sputum samples using shotgun metagenomics on a benchtop sequencer.
Doughty EL, Sergeant MJ, Adetifa I, Antonio M, Pallen MJ. Doughty EL, et al. PeerJ. 2014 Sep 23;2:e585. doi: 10.7717/peerj.585. eCollection 2014. PeerJ. 2014. PMID: 25279265 Free PMC article. - Rapid draft sequencing and real-time nanopore sequencing in a hospital outbreak of Salmonella.
Quick J, Ashton P, Calus S, Chatt C, Gossain S, Hawker J, Nair S, Neal K, Nye K, Peters T, De Pinna E, Robinson E, Struthers K, Webber M, Catto A, Dallman TJ, Hawkey P, Loman NJ. Quick J, et al. Genome Biol. 2015 May 30;16:114. doi: 10.1186/s13059-015-0677-2. Genome Biol. 2015. PMID: 26025440 Free PMC article. - Full genome SNP-based phylogenetic analysis reveals the origin and global spread of Brucella melitensis.
Tan KK, Tan YC, Chang LY, Lee KW, Nore SS, Yee WY, Mat Isa MN, Jafar FL, Hoh CC, AbuBakar S. Tan KK, et al. BMC Genomics. 2015 Feb 18;16(1):93. doi: 10.1186/s12864-015-1294-x. BMC Genomics. 2015. PMID: 25888205 Free PMC article. - Evolutionary history and current distribution of the West Mediterranean lineage of Brucella melitensis in Italy.
Janowicz A, De Massis F, Zilli K, Ancora M, Tittarelli M, Sacchini F, Di Giannatale E, Sahl JW, Foster JT, Garofolo G. Janowicz A, et al. Microb Genom. 2020 Nov;6(11):mgen000446. doi: 10.1099/mgen.0.000446. Epub 2020 Oct 8. Microb Genom. 2020. PMID: 33030422 Free PMC article.
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous