OAS proteins and cGAS: unifying concepts in sensing and responding to cytosolic nucleic acids - PubMed (original) (raw)
Review
OAS proteins and cGAS: unifying concepts in sensing and responding to cytosolic nucleic acids
Veit Hornung et al. Nat Rev Immunol. 2014 Aug.
Abstract
Recent discoveries in the field of innate immunity have highlighted the existence of a family of nucleic acid-sensing proteins that have similar structural and functional properties. These include the well-known oligoadenylate synthase (OAS) family proteins and the recently identified OAS homologue cyclic GMP-AMP (cGAMP) synthase (cGAS). The OAS proteins and cGAS are template-independent nucleotidyltransferases that, once activated by double-stranded nucleic acids in the cytosol, produce unique classes of 2'-5'-linked second messenger molecules, which - through distinct mechanisms - have crucial antiviral functions. 2'-5'-linked oligoadenylates limit viral propagation through the activation of the enzyme RNase L, which degrades host and viral RNA, and 2'-5'-linked cGAMP activates downstream signalling pathways to induce de novo antiviral gene expression. In this Progress article, we describe the striking functional and structural similarities between OAS proteins and cGAS, and highlight their roles in antiviral immunity.
Conflict of interest statement
The authors declare no competing financial interests.
Figures
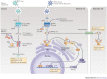
Figure 1. Simplified schematic comparison of the OAS1–RNase L and cGAS–STING axes in innate immune signalling and antiviral defence.
Upon double-stranded RNA (dsRNA) binding, oligoadenylate synthase (OAS) enzymes undergo a conformational switch, which results in their catalytic activity — that is, the synthesis of 2′–5′-linked oligoadenylates using ATP as a substrate. 2′–5′-linked oligoadenylates subsequently act as second messenger molecules by activating the latent endoribonuclease RNase L in the cytoplasm. RNase L then forms a crossed dimer and degrades RNA that is of both cellular and viral origin, leading to the inhibition of viral propagation. On the other hand, cyclic GMP–AMP (cGAMP) synthase (cGAS) is activated by cytosolic B-form dsDNA to synthesize the non-canonical cyclic dinucleotide (CDN) cGAMP(2′–5′) as its second messenger molecule (using the substrates ATP and GTP). cGAMP(2′–5′) binds to and activates the endoplasmic reticulum (ER)-resident receptor stimulator of interferon genes (STING), which subsequently translocates to a perinuclear Golgi compartment where it obtains its signalling-competent state. This results in the activation of transcription factors that initiate antiviral and pro-inflammatory gene expression. At the same time, cGAMP(2′–5′) can also diffuse through gap junctions to initiate antiviral activity in bystander cells. In addition to its role in sensing the endogenous second messenger molecule cGAMP(2′–5′), STING responds to exogenous CDNs that are derived from prokaryotes (not shown). IκB, inhibitor of NF-κB; IKK, IκB kinase complex; IRF3, interferon-regulatory factor 3; NF-κB, nuclear factor-κB; TBK1, TANK-binding kinase 1. PowerPoint slide
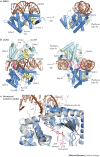
Figure 2. Structures of OAS1 and cGAS.
a | Two views of 2′–5′-oligoadenylate synthase 1 (OAS1) in complex with double-stranded RNA (dsRNA). b | Two views of the cyclic GMP–AMP (cGAMP) synthase (cGAS) dimer (light and dark blue) in complex with dsDNA. c | Close up comparison of cGAS in the apo form (grey) and DNA-bound form (blue). DNA binds to the platform at site A and induces a structural switch in the spine helix (indicated by arrows) and active site loops, thereby facilitating ATP and GTP binding and catalysis. ZT, zinc thumb. PowerPoint slide
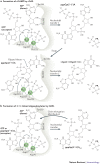
Figure 3. Unified mechanism of nucleotidyl transfer by cGAS and OAS1 proteins.
a | The mechanism for two-step nucleotidyl transfer in the formation of cyclic GMP–AMP (cGAMP) by cGAMP synthase (cGAS). Selected catalytically important residues, as well as two magnesium ions, are indicated. In particular, Glu225 and Asp227 bind the two magnesium ions that are crucial to orient and activate the donor triphosphate moiety. Asp319 polarizes the attacking 2′-OH, and Tyr436 and Glu383 bind base and ribose, respectively. After the first catalytic step, the pppGp(2′–5′)A intermediate needs to dissociate and rebind in reverse order (swap) for the second catalytic step. Human cGAS numbering is used for the amino acid residues shown. b | Nucleotidyl transfer 1 by oligoadenylate synthase 1 (OAS1) is similar to that of cGAS. Instead of a nucleotide swap, as observed in cGAS, the product of nucleotidyl transfer 1 rebinds to the acceptor site only. In the beginning, the acceptor is ATP (in ATP acceptor, 'R' denotes diphosphate group), whereas in subsequent steps, during chain elongation, the acceptor is pppAp(2′–5′)A and longer 2′–5′-linked oligoadenylates ('R' denotes pppAp(2′–5′)A and longer chains). Human OAS1 numbering is used for the amino acid residues shown. PowerPoint slide
Similar articles
- Cyclic GMP-AMP as an Endogenous Second Messenger in Innate Immune Signaling by Cytosolic DNA.
Kato K, Omura H, Ishitani R, Nureki O. Kato K, et al. Annu Rev Biochem. 2017 Jun 20;86:541-566. doi: 10.1146/annurev-biochem-061516-044813. Epub 2017 Apr 7. Annu Rev Biochem. 2017. PMID: 28399655 Review. - Origin and development of oligoadenylate synthetase immune system.
Hu J, Wang X, Xing Y, Rong E, Ning M, Smith J, Huang Y. Hu J, et al. BMC Evol Biol. 2018 Dec 27;18(1):201. doi: 10.1186/s12862-018-1315-x. BMC Evol Biol. 2018. PMID: 30587119 Free PMC article. - The Role of Phosphodiesterase 12 (PDE12) as a Negative Regulator of the Innate Immune Response and the Discovery of Antiviral Inhibitors.
Wood ER, Bledsoe R, Chai J, Daka P, Deng H, Ding Y, Harris-Gurley S, Kryn LH, Nartey E, Nichols J, Nolte RT, Prabhu N, Rise C, Sheahan T, Shotwell JB, Smith D, Tai V, Taylor JD, Tomberlin G, Wang L, Wisely B, You S, Xia B, Dickson H. Wood ER, et al. J Biol Chem. 2015 Aug 7;290(32):19681-96. doi: 10.1074/jbc.M115.653113. Epub 2015 Jun 8. J Biol Chem. 2015. PMID: 26055709 Free PMC article. - The Association of OASL and Type I Interferons in the Pathogenesis and Survival of Intracellular Replicating Bacterial Species.
Leisching G, Wiid I, Baker B. Leisching G, et al. Front Cell Infect Microbiol. 2017 May 19;7:196. doi: 10.3389/fcimb.2017.00196. eCollection 2017. Front Cell Infect Microbiol. 2017. PMID: 28580319 Free PMC article. Review. - The Many Faces of Oligoadenylate Synthetases.
Sarkar SN, Harioudh MK, Shao L, Perez J, Ghosh A. Sarkar SN, et al. J Interferon Cytokine Res. 2023 Nov;43(11):487-494. doi: 10.1089/jir.2023.0098. Epub 2023 Sep 25. J Interferon Cytokine Res. 2023. PMID: 37751211 Free PMC article. Review.
Cited by
- Synthetic Cell Lines for Inducible Packaging of Influenza A Virus.
Phan T, Ye Q, Stach C, Lin YC, Cao H, Bowen A, Langlois RA, Hu WS. Phan T, et al. ACS Synth Biol. 2024 Feb 16;13(2):546-557. doi: 10.1021/acssynbio.3c00526. Epub 2024 Jan 23. ACS Synth Biol. 2024. PMID: 38259154 Free PMC article. - Protein N-myristoylation: functions and mechanisms in control of innate immunity.
Wang B, Dai T, Sun W, Wei Y, Ren J, Zhang L, Zhang M, Zhou F. Wang B, et al. Cell Mol Immunol. 2021 Apr;18(4):878-888. doi: 10.1038/s41423-021-00663-2. Epub 2021 Mar 17. Cell Mol Immunol. 2021. PMID: 33731917 Free PMC article. Review. - At the Crossroads of the cGAS-cGAMP-STING Pathway and the DNA Damage Response: Implications for Cancer Progression and Treatment.
Korneenko TV, Pestov NB, Nevzorov IA, Daks AA, Trachuk KN, Solopova ON, Barlev NA. Korneenko TV, et al. Pharmaceuticals (Basel). 2023 Dec 1;16(12):1675. doi: 10.3390/ph16121675. Pharmaceuticals (Basel). 2023. PMID: 38139802 Free PMC article. Review. - Enterococcus casseliflavus KB1733 Isolated from a Traditional Japanese Pickle Induces Interferon-Lambda Production in Human Intestinal Epithelial Cells.
Satomi S, Kokubu D, Inoue T, Sugiyama M, Mizokami M, Suzuki S, Murata K. Satomi S, et al. Microorganisms. 2022 Apr 15;10(4):827. doi: 10.3390/microorganisms10040827. Microorganisms. 2022. PMID: 35456876 Free PMC article. - The Evolutionary Dance between Innate Host Antiviral Pathways and SARS-CoV-2.
Aliyari SR, Quanquin N, Pernet O, Zhang S, Wang L, Cheng G. Aliyari SR, et al. Pathogens. 2022 May 3;11(5):538. doi: 10.3390/pathogens11050538. Pathogens. 2022. PMID: 35631059 Free PMC article. Review.
References
- Medzhitov R. Recognition of microorganisms and activation of the immune response. Nature. 2007;449:819–826. - PubMed
- Chebath J, Benech P, Revel M, Vigneron M. Constitutive expression of (2′–5′) oligo A synthetase confers resistance to picornavirus infection. Nature. 1987;330:587–588. - PubMed
- Melchjorsen J, et al. Differential regulation of the OASL and OAS1 genes in response to viral infections. J. Interferon Cytokine Res. 2009;29:199–207. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources