Origin of giant viruses from smaller DNA viruses not from a fourth domain of cellular life - PubMed (original) (raw)
Origin of giant viruses from smaller DNA viruses not from a fourth domain of cellular life
Natalya Yutin et al. Virology. 2014 Oct.
Abstract
The numerous and diverse eukaryotic viruses with large double-stranded DNA genomes that at least partially reproduce in the cytoplasm of infected cells apparently evolved from a single virus ancestor. This major group of viruses is known as Nucleocytoplasmic Large DNA Viruses (NCLDV) or the proposed order Megavirales. Among the "Megavirales", there are three groups of giant viruses with genomes exceeding 500kb, namely Mimiviruses, Pithoviruses, and Pandoraviruses that hold the current record of viral genome size, about 2.5Mb. Phylogenetic analysis of conserved, ancestral NLCDV genes clearly shows that these three groups of giant viruses have three distinct origins within the "Megavirales". The Mimiviruses constitute a distinct family that is distantly related to Phycodnaviridae, Pandoraviruses originate from a common ancestor with Coccolithoviruses within the Phycodnaviridae family, and Pithoviruses are related to Iridoviridae and Marseilleviridae. Maximum likelihood reconstruction of gene gain and loss events during the evolution of the "Megavirales" indicates that each group of giant viruses evolved from viruses with substantially smaller and simpler gene repertoires. Initial phylogenetic analysis of universal genes, such as translation system components, encoded by some giant viruses, in particular Mimiviruses, has led to the hypothesis that giant viruses descend from a fourth, probably extinct domain of cellular life. The results of our comprehensive phylogenomic analysis of giant viruses refute the fourth domain hypothesis and instead indicate that the universal genes have been independently acquired by different giant viruses from their eukaryotic hosts.
Keywords: Domains of cellular life; Genome evolution; Giant viruses; Phylogenomics.
Published by Elsevier Inc.
Figures
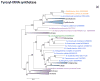
Figure 1. Phylogenies of the large subunits of DNA-dependent RNA polymerase
(a) Subunit a (b) Subunit b Support values represent expected-likelihood weights of 1,000 local rearrangements; branches with support less than 50 were collapsed. “Megavirales” sequences are highlighted in orange, eukaryotic sequences in blue, archaeal sequences in purple. OLPG: Organic Lake phycodnavirus – Phaeocystic globosa virus clade.
Figure 2. Phylogenies of aminoacyl-tRNA synthetases encoded by giant viruses
(a) Tyrosyl-tRNA synthetase (b) Arginyl-tRNA synthetase (c) Aspartyl/asparaginyl-tRNA synthetases (d) Cysteinyl-tRNA synthetase (e) Isoleucyl-tRNA synthetase (f) Methionyl-tRNA synthetase (g) Tryptophanyl-tRNA synthetase Support values represent expected-likelihood weights of 1,000 local rearrangements; branches with support less than 50 were collapsed. “Megavirales” sequences are highlighted in orange, eukaryotic sequences in blue, bacterial sequences in green, archaeal sequences in purple. Taxa abbreviations: Ac, Crenarchaeota; Ae, Euryarchaeota; Az, unclassified Archaea; Ba, Actinobacteria; Bb, Bacteroidetes/Chlorobi group; Bc, Cyanobacteria; Bi, Acidobacteria; Bp, Proteobacteria; Bs, Spirochaetes; Bv, Chlamydiae/Verrucomicrobia group; E2, Fornicata; E7, Rhodophyta; E8, stramenopiles; E9, Viridiplantae; Ea, Amoebozoa; Ec, Alveolata; Eh, Cryptophyta; Ek, Euglenozoa; El, Opisthokonta; Eq, Heterolobosea; Ew, Parabasalidea.
Figure 2. Phylogenies of aminoacyl-tRNA synthetases encoded by giant viruses
(a) Tyrosyl-tRNA synthetase (b) Arginyl-tRNA synthetase (c) Aspartyl/asparaginyl-tRNA synthetases (d) Cysteinyl-tRNA synthetase (e) Isoleucyl-tRNA synthetase (f) Methionyl-tRNA synthetase (g) Tryptophanyl-tRNA synthetase Support values represent expected-likelihood weights of 1,000 local rearrangements; branches with support less than 50 were collapsed. “Megavirales” sequences are highlighted in orange, eukaryotic sequences in blue, bacterial sequences in green, archaeal sequences in purple. Taxa abbreviations: Ac, Crenarchaeota; Ae, Euryarchaeota; Az, unclassified Archaea; Ba, Actinobacteria; Bb, Bacteroidetes/Chlorobi group; Bc, Cyanobacteria; Bi, Acidobacteria; Bp, Proteobacteria; Bs, Spirochaetes; Bv, Chlamydiae/Verrucomicrobia group; E2, Fornicata; E7, Rhodophyta; E8, stramenopiles; E9, Viridiplantae; Ea, Amoebozoa; Ec, Alveolata; Eh, Cryptophyta; Ek, Euglenozoa; El, Opisthokonta; Eq, Heterolobosea; Ew, Parabasalidea.
Figure 2. Phylogenies of aminoacyl-tRNA synthetases encoded by giant viruses
(a) Tyrosyl-tRNA synthetase (b) Arginyl-tRNA synthetase (c) Aspartyl/asparaginyl-tRNA synthetases (d) Cysteinyl-tRNA synthetase (e) Isoleucyl-tRNA synthetase (f) Methionyl-tRNA synthetase (g) Tryptophanyl-tRNA synthetase Support values represent expected-likelihood weights of 1,000 local rearrangements; branches with support less than 50 were collapsed. “Megavirales” sequences are highlighted in orange, eukaryotic sequences in blue, bacterial sequences in green, archaeal sequences in purple. Taxa abbreviations: Ac, Crenarchaeota; Ae, Euryarchaeota; Az, unclassified Archaea; Ba, Actinobacteria; Bb, Bacteroidetes/Chlorobi group; Bc, Cyanobacteria; Bi, Acidobacteria; Bp, Proteobacteria; Bs, Spirochaetes; Bv, Chlamydiae/Verrucomicrobia group; E2, Fornicata; E7, Rhodophyta; E8, stramenopiles; E9, Viridiplantae; Ea, Amoebozoa; Ec, Alveolata; Eh, Cryptophyta; Ek, Euglenozoa; El, Opisthokonta; Eq, Heterolobosea; Ew, Parabasalidea.
Figure 2. Phylogenies of aminoacyl-tRNA synthetases encoded by giant viruses
(a) Tyrosyl-tRNA synthetase (b) Arginyl-tRNA synthetase (c) Aspartyl/asparaginyl-tRNA synthetases (d) Cysteinyl-tRNA synthetase (e) Isoleucyl-tRNA synthetase (f) Methionyl-tRNA synthetase (g) Tryptophanyl-tRNA synthetase Support values represent expected-likelihood weights of 1,000 local rearrangements; branches with support less than 50 were collapsed. “Megavirales” sequences are highlighted in orange, eukaryotic sequences in blue, bacterial sequences in green, archaeal sequences in purple. Taxa abbreviations: Ac, Crenarchaeota; Ae, Euryarchaeota; Az, unclassified Archaea; Ba, Actinobacteria; Bb, Bacteroidetes/Chlorobi group; Bc, Cyanobacteria; Bi, Acidobacteria; Bp, Proteobacteria; Bs, Spirochaetes; Bv, Chlamydiae/Verrucomicrobia group; E2, Fornicata; E7, Rhodophyta; E8, stramenopiles; E9, Viridiplantae; Ea, Amoebozoa; Ec, Alveolata; Eh, Cryptophyta; Ek, Euglenozoa; El, Opisthokonta; Eq, Heterolobosea; Ew, Parabasalidea.
Figure 2. Phylogenies of aminoacyl-tRNA synthetases encoded by giant viruses
(a) Tyrosyl-tRNA synthetase (b) Arginyl-tRNA synthetase (c) Aspartyl/asparaginyl-tRNA synthetases (d) Cysteinyl-tRNA synthetase (e) Isoleucyl-tRNA synthetase (f) Methionyl-tRNA synthetase (g) Tryptophanyl-tRNA synthetase Support values represent expected-likelihood weights of 1,000 local rearrangements; branches with support less than 50 were collapsed. “Megavirales” sequences are highlighted in orange, eukaryotic sequences in blue, bacterial sequences in green, archaeal sequences in purple. Taxa abbreviations: Ac, Crenarchaeota; Ae, Euryarchaeota; Az, unclassified Archaea; Ba, Actinobacteria; Bb, Bacteroidetes/Chlorobi group; Bc, Cyanobacteria; Bi, Acidobacteria; Bp, Proteobacteria; Bs, Spirochaetes; Bv, Chlamydiae/Verrucomicrobia group; E2, Fornicata; E7, Rhodophyta; E8, stramenopiles; E9, Viridiplantae; Ea, Amoebozoa; Ec, Alveolata; Eh, Cryptophyta; Ek, Euglenozoa; El, Opisthokonta; Eq, Heterolobosea; Ew, Parabasalidea.
Figure 2. Phylogenies of aminoacyl-tRNA synthetases encoded by giant viruses
(a) Tyrosyl-tRNA synthetase (b) Arginyl-tRNA synthetase (c) Aspartyl/asparaginyl-tRNA synthetases (d) Cysteinyl-tRNA synthetase (e) Isoleucyl-tRNA synthetase (f) Methionyl-tRNA synthetase (g) Tryptophanyl-tRNA synthetase Support values represent expected-likelihood weights of 1,000 local rearrangements; branches with support less than 50 were collapsed. “Megavirales” sequences are highlighted in orange, eukaryotic sequences in blue, bacterial sequences in green, archaeal sequences in purple. Taxa abbreviations: Ac, Crenarchaeota; Ae, Euryarchaeota; Az, unclassified Archaea; Ba, Actinobacteria; Bb, Bacteroidetes/Chlorobi group; Bc, Cyanobacteria; Bi, Acidobacteria; Bp, Proteobacteria; Bs, Spirochaetes; Bv, Chlamydiae/Verrucomicrobia group; E2, Fornicata; E7, Rhodophyta; E8, stramenopiles; E9, Viridiplantae; Ea, Amoebozoa; Ec, Alveolata; Eh, Cryptophyta; Ek, Euglenozoa; El, Opisthokonta; Eq, Heterolobosea; Ew, Parabasalidea.
Figure 2. Phylogenies of aminoacyl-tRNA synthetases encoded by giant viruses
(a) Tyrosyl-tRNA synthetase (b) Arginyl-tRNA synthetase (c) Aspartyl/asparaginyl-tRNA synthetases (d) Cysteinyl-tRNA synthetase (e) Isoleucyl-tRNA synthetase (f) Methionyl-tRNA synthetase (g) Tryptophanyl-tRNA synthetase Support values represent expected-likelihood weights of 1,000 local rearrangements; branches with support less than 50 were collapsed. “Megavirales” sequences are highlighted in orange, eukaryotic sequences in blue, bacterial sequences in green, archaeal sequences in purple. Taxa abbreviations: Ac, Crenarchaeota; Ae, Euryarchaeota; Az, unclassified Archaea; Ba, Actinobacteria; Bb, Bacteroidetes/Chlorobi group; Bc, Cyanobacteria; Bi, Acidobacteria; Bp, Proteobacteria; Bs, Spirochaetes; Bv, Chlamydiae/Verrucomicrobia group; E2, Fornicata; E7, Rhodophyta; E8, stramenopiles; E9, Viridiplantae; Ea, Amoebozoa; Ec, Alveolata; Eh, Cryptophyta; Ek, Euglenozoa; El, Opisthokonta; Eq, Heterolobosea; Ew, Parabasalidea.
Figure 3. Phylogenies of translation encoded in “Megavirales” genomes
(a) elongation factor EF-1alpha (b) initiation factor eIF-1 (SUI1) (c) initiation factor eIF-2beta (d) initiation factor eIF-4A (e) initiation factor SUA5 (f) peptide chain release factor eRF1 Support values represent expected-likelihood weights of 1,000 local rearrangements; branches with support less than 50 were collapsed. “Megavirales” sequences are highlighted in orange, eukaryotic sequences in blue, archaeal sequences in purple. Taxa abbreviations: Ae, Euryarchaeota; Ba, Actinobacteria; Bf, Firmicutes; Bj, Tenericutes; Bp, Proteobacteria; E2, Fornicata; E7, Rhodophyta; E9, Viridiplantae; Ea, Amoebozoa; Ec, Alveolata; Eh, Cryptophyta; Ek, Euglenozoa; El, Opisthokonta; Ep, Haptophyceae; Eq, Heterolobosea; Ew, Parabasalidea.
Figure 3. Phylogenies of translation encoded in “Megavirales” genomes
(a) elongation factor EF-1alpha (b) initiation factor eIF-1 (SUI1) (c) initiation factor eIF-2beta (d) initiation factor eIF-4A (e) initiation factor SUA5 (f) peptide chain release factor eRF1 Support values represent expected-likelihood weights of 1,000 local rearrangements; branches with support less than 50 were collapsed. “Megavirales” sequences are highlighted in orange, eukaryotic sequences in blue, archaeal sequences in purple. Taxa abbreviations: Ae, Euryarchaeota; Ba, Actinobacteria; Bf, Firmicutes; Bj, Tenericutes; Bp, Proteobacteria; E2, Fornicata; E7, Rhodophyta; E9, Viridiplantae; Ea, Amoebozoa; Ec, Alveolata; Eh, Cryptophyta; Ek, Euglenozoa; El, Opisthokonta; Ep, Haptophyceae; Eq, Heterolobosea; Ew, Parabasalidea.
Figure 3. Phylogenies of translation encoded in “Megavirales” genomes
(a) elongation factor EF-1alpha (b) initiation factor eIF-1 (SUI1) (c) initiation factor eIF-2beta (d) initiation factor eIF-4A (e) initiation factor SUA5 (f) peptide chain release factor eRF1 Support values represent expected-likelihood weights of 1,000 local rearrangements; branches with support less than 50 were collapsed. “Megavirales” sequences are highlighted in orange, eukaryotic sequences in blue, archaeal sequences in purple. Taxa abbreviations: Ae, Euryarchaeota; Ba, Actinobacteria; Bf, Firmicutes; Bj, Tenericutes; Bp, Proteobacteria; E2, Fornicata; E7, Rhodophyta; E9, Viridiplantae; Ea, Amoebozoa; Ec, Alveolata; Eh, Cryptophyta; Ek, Euglenozoa; El, Opisthokonta; Ep, Haptophyceae; Eq, Heterolobosea; Ew, Parabasalidea.
Figure 3. Phylogenies of translation encoded in “Megavirales” genomes
(a) elongation factor EF-1alpha (b) initiation factor eIF-1 (SUI1) (c) initiation factor eIF-2beta (d) initiation factor eIF-4A (e) initiation factor SUA5 (f) peptide chain release factor eRF1 Support values represent expected-likelihood weights of 1,000 local rearrangements; branches with support less than 50 were collapsed. “Megavirales” sequences are highlighted in orange, eukaryotic sequences in blue, archaeal sequences in purple. Taxa abbreviations: Ae, Euryarchaeota; Ba, Actinobacteria; Bf, Firmicutes; Bj, Tenericutes; Bp, Proteobacteria; E2, Fornicata; E7, Rhodophyta; E9, Viridiplantae; Ea, Amoebozoa; Ec, Alveolata; Eh, Cryptophyta; Ek, Euglenozoa; El, Opisthokonta; Ep, Haptophyceae; Eq, Heterolobosea; Ew, Parabasalidea.
Figure 3. Phylogenies of translation encoded in “Megavirales” genomes
(a) elongation factor EF-1alpha (b) initiation factor eIF-1 (SUI1) (c) initiation factor eIF-2beta (d) initiation factor eIF-4A (e) initiation factor SUA5 (f) peptide chain release factor eRF1 Support values represent expected-likelihood weights of 1,000 local rearrangements; branches with support less than 50 were collapsed. “Megavirales” sequences are highlighted in orange, eukaryotic sequences in blue, archaeal sequences in purple. Taxa abbreviations: Ae, Euryarchaeota; Ba, Actinobacteria; Bf, Firmicutes; Bj, Tenericutes; Bp, Proteobacteria; E2, Fornicata; E7, Rhodophyta; E9, Viridiplantae; Ea, Amoebozoa; Ec, Alveolata; Eh, Cryptophyta; Ek, Euglenozoa; El, Opisthokonta; Ep, Haptophyceae; Eq, Heterolobosea; Ew, Parabasalidea.
Figure 3. Phylogenies of translation encoded in “Megavirales” genomes
(a) elongation factor EF-1alpha (b) initiation factor eIF-1 (SUI1) (c) initiation factor eIF-2beta (d) initiation factor eIF-4A (e) initiation factor SUA5 (f) peptide chain release factor eRF1 Support values represent expected-likelihood weights of 1,000 local rearrangements; branches with support less than 50 were collapsed. “Megavirales” sequences are highlighted in orange, eukaryotic sequences in blue, archaeal sequences in purple. Taxa abbreviations: Ae, Euryarchaeota; Ba, Actinobacteria; Bf, Firmicutes; Bj, Tenericutes; Bp, Proteobacteria; E2, Fornicata; E7, Rhodophyta; E9, Viridiplantae; Ea, Amoebozoa; Ec, Alveolata; Eh, Cryptophyta; Ek, Euglenozoa; El, Opisthokonta; Ep, Haptophyceae; Eq, Heterolobosea; Ew, Parabasalidea.
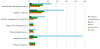
Figure 4
Phylogenomic breakdown of giant virus genes
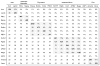
Figure 5. Relationships between the gene contents of giant viruses and their smaller relatives
(a) matrix of shared genes Lower left: number of shared gene families. Upper right: Jaccard similarity of gene complements. Diagonal: number of annotated genes in the genome. Intra-family comparisons are shaded. (b) tree of gene contents Bold lines indicate branches with high (>70%) bootstrap support; thin lines indicate branches with low bootstrap support. Branches that disagree with the tree, reconstructed with the universal core genes, are highlighted in red (except the poorly resolved branches inside the Poxviridae), dashed lines indicate the relationships expected from the core phylogeny.
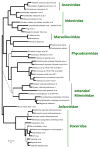
Figure 6. Phylogenetic tree of the (nearly) universal core genes of the “Megavirales” and reconstruction of gene gain and loss
Numbers above the branch indicate the estimated number of NCVOG families (plus the number of singletons for extant genomes) at the end of the branch. Numbers below the branch indicate the estimated number of gained and lost NCVOG families (plus the number of acquired singletons for extant genomes). Dashed lines extend the branches The estimated number of “Megavirales” ancestral families is indicated in a circle at the tree root.
Figure 6. Phylogenetic tree of the (nearly) universal core genes of the “Megavirales” and reconstruction of gene gain and loss
Numbers above the branch indicate the estimated number of NCVOG families (plus the number of singletons for extant genomes) at the end of the branch. Numbers below the branch indicate the estimated number of gained and lost NCVOG families (plus the number of acquired singletons for extant genomes). Dashed lines extend the branches The estimated number of “Megavirales” ancestral families is indicated in a circle at the tree root.
Similar articles
- Evolution of the Large Nucleocytoplasmic DNA Viruses of Eukaryotes and Convergent Origins of Viral Gigantism.
Koonin EV, Yutin N. Koonin EV, et al. Adv Virus Res. 2019;103:167-202. doi: 10.1016/bs.aivir.2018.09.002. Epub 2018 Nov 10. Adv Virus Res. 2019. PMID: 30635076 Review. - Virus Genomes from Deep Sea Sediments Expand the Ocean Megavirome and Support Independent Origins of Viral Gigantism.
Bäckström D, Yutin N, Jørgensen SL, Dharamshi J, Homa F, Zaremba-Niedwiedzka K, Spang A, Wolf YI, Koonin EV, Ettema TJG. Bäckström D, et al. mBio. 2019 Mar 5;10(2):e02497-18. doi: 10.1128/mBio.02497-18. mBio. 2019. PMID: 30837339 Free PMC article. - Eukaryotic large nucleo-cytoplasmic DNA viruses: clusters of orthologous genes and reconstruction of viral genome evolution.
Yutin N, Wolf YI, Raoult D, Koonin EV. Yutin N, et al. Virol J. 2009 Dec 17;6:223. doi: 10.1186/1743-422X-6-223. Virol J. 2009. PMID: 20017929 Free PMC article. - Evolution of double-stranded DNA viruses of eukaryotes: from bacteriophages to transposons to giant viruses.
Koonin EV, Krupovic M, Yutin N. Koonin EV, et al. Ann N Y Acad Sci. 2015 Apr;1341(1):10-24. doi: 10.1111/nyas.12728. Epub 2015 Feb 27. Ann N Y Acad Sci. 2015. PMID: 25727355 Free PMC article. Review. - "Megavirales", a proposed new order for eukaryotic nucleocytoplasmic large DNA viruses.
Colson P, De Lamballerie X, Yutin N, Asgari S, Bigot Y, Bideshi DK, Cheng XW, Federici BA, Van Etten JL, Koonin EV, La Scola B, Raoult D. Colson P, et al. Arch Virol. 2013 Dec;158(12):2517-21. doi: 10.1007/s00705-013-1768-6. Epub 2013 Jun 29. Arch Virol. 2013. PMID: 23812617 Free PMC article.
Cited by
- Structure-Based Deep Mining Reveals First-Time Annotations for 46 Percent of the Dark Annotation Space of the 9,671-Member Superproteome of the Nucleocytoplasmic Large DNA Viruses.
Mirzakhanyan Y, Gershon PD. Mirzakhanyan Y, et al. J Virol. 2020 Nov 23;94(24):e00854-20. doi: 10.1128/JVI.00854-20. Print 2020 Nov 23. J Virol. 2020. PMID: 32999026 Free PMC article. - Genomes of six viruses that infect Asgard archaea from deep-sea sediments.
Rambo IM, Langwig MV, Leão P, De Anda V, Baker BJ. Rambo IM, et al. Nat Microbiol. 2022 Jul;7(7):953-961. doi: 10.1038/s41564-022-01150-8. Epub 2022 Jun 27. Nat Microbiol. 2022. PMID: 35760837 - Medusavirus, a Novel Large DNA Virus Discovered from Hot Spring Water.
Yoshikawa G, Blanc-Mathieu R, Song C, Kayama Y, Mochizuki T, Murata K, Ogata H, Takemura M. Yoshikawa G, et al. J Virol. 2019 Apr 3;93(8):e02130-18. doi: 10.1128/JVI.02130-18. Print 2019 Apr 15. J Virol. 2019. PMID: 30728258 Free PMC article. - The number of genes encoding repeat domain-containing proteins positively correlates with genome size in amoebal giant viruses.
Shukla A, Chatterjee A, Kondabagil K. Shukla A, et al. Virus Evol. 2018 Jan 3;4(1):vex039. doi: 10.1093/ve/vex039. eCollection 2018 Jan. Virus Evol. 2018. PMID: 29308275 Free PMC article. - Minimizing the Anticodon-Recognized Loop of Methanococcus jannaschii Tyrosyl-tRNA Synthetase to Improve the Efficiency of Incorporating Noncanonical Amino Acids.
Hu Z, Liang J, Su T, Zhang D, Li H, Gao X, Yao W, Song X. Hu Z, et al. Biomolecules. 2023 Mar 28;13(4):610. doi: 10.3390/biom13040610. Biomolecules. 2023. PMID: 37189358 Free PMC article.
References
- Brown JR, Douady CJ, Italia MJ, Marshall WE, Stanhope MJ. Universal trees based on large combined protein sequence data sets. Nat Genet. 2001;28(3):281–5. - PubMed
- Ciccarelli FD, Doerks T, von Mering C, Creevey CJ, Snel B, Bork P. Toward automatic reconstruction of a highly resolved tree of life. Science. 2006;311(5765):1283–7. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources