A novel stable isotope labelling assisted workflow for improved untargeted LC-HRMS based metabolomics research - PubMed (original) (raw)
doi: 10.1007/s11306-013-0611-0. Epub 2013 Dec 4.
Bernhard Kluger 1, Marc Lemmens 1, Gerhard Adam 2, Gerlinde Wiesenberger 2, Valentina Maschietto 3, Adriano Marocco 3, Joseph Strauss 2 4, Stephan Bödi 2, Gerhard G Thallinger 5 6, Rudolf Krska 1, Rainer Schuhmacher 1
Affiliations
- PMID: 25057268
- PMCID: PMC4098048
- DOI: 10.1007/s11306-013-0611-0
A novel stable isotope labelling assisted workflow for improved untargeted LC-HRMS based metabolomics research
Christoph Bueschl et al. Metabolomics. 2014.
Abstract
Many untargeted LC-ESI-HRMS based metabolomics studies are still hampered by the large proportion of non-biological sample derived signals included in the generated raw data. Here, a novel, powerful stable isotope labelling (SIL)-based metabolomics workflow is presented, which facilitates global metabolome extraction, improved metabolite annotation and metabolome wide internal standardisation (IS). The general concept is exemplified with two different cultivation variants, (1) co-cultivation of the plant pathogenic fungi Fusarium graminearum on non-labelled and highly 13C enriched culture medium and (2) experimental cultivation under native conditions and use of globally U-13C labelled biological reference samples as exemplified with maize and wheat. Subsequent to LC-HRMS analysis of mixtures of labelled and non-labelled samples, two-dimensional data filtering of SIL specific isotopic patterns is performed to better extract truly biological derived signals together with the corresponding number of carbon atoms of each metabolite ion. Finally, feature pairs are convoluted to feature groups each representing a single metabolite. Moreover, the correction of unequal matrix effects in different sample types and the improvement of relative metabolite quantification with metabolome wide IS are demonstrated for the F. graminearum experiment. Data processing employing the presented workflow revealed about 300 SIL derived feature pairs corresponding to 87-135 metabolites in F. graminearum samples and around 800 feature pairs corresponding to roughly 350 metabolites in wheat samples. SIL assisted IS, by the use of globally U-13C labelled biological samples, reduced the median CV value from 7.1 to 3.6 % for technical replicates and from 15.1 to 10.8 % for biological replicates in the respective F. graminearum samples.
Keywords: 13C-labelling; Fusarium; Internal standardisation; Maize; Metabolomics; Wheat.
Figures
Fig. 1
Overview of the proposed SIL assisted workflow for native and U-13C co-cultivation (variant A) and native cultivation and use of U-13C reference metabolome (variant B) [figure-width: 174 mm]
Fig. 2
3D representation of a selected F. graminearum aggregate sample analysed with LC–HRMS. Chromatogram of the unprocessed, centroided (a) and the processed (b) with only the SIL derived MS signals are shown. The 3D representation in c shows a zoomed section of the unprocessed datafile (a) illustrating the labelling specific isotopic pattern for three different ion species (M denotes the monoisotopic 12C metabolite and _M_′ denotes the U-13C labelled metabolite) of a metabolite with the neutral, monoisotopic mass of 624.3827 u and n C = 30 carbon atoms. 3D representations were created with TOPPView (Sturm and Kohlbacher , v. 1.10) [figure-width: 174 mm]
Fig. 3
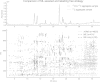
a Illustration of an overlay of full scan LC–HRMS total ion current chromatograms obtained for two F. graminearum aggregate samples. Red Non-labelled 12C and U-13C culture filtrate mixed 1:1 (v/v); grey Non-labelled filtrate mixed 1:1 with fungal growth medium. b 2D plot of detected LC–HRMS features (all dots). Grey symbols indicate all features found with XCMS processing. Red symbols represent monoisotopic 12C features found by both XCMS and the presented workflow (variant A, Fig. 1). Monoisotopic 12C features found by the labelling assisted approach only are marked in blue. Features with a retention time >30 min are mainly detected by XCMS. Due to the higher strength of the eluent, predominantly impurities of non-biological origin such as polymers and apolar compounds are displaced from the stationary phase [figure-width: 174 mm]
Fig. 4
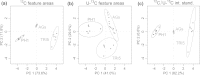
Three PCA scores plots derived from consistently extracted feature pairs of three sample types: F. graminearum samples PH-1, _tri5_Δ and aggregate samples (AGs). For all three PCAs the exactly same set of feature pairs was used, however different intensity values (peak areas) were taken for each feature pair. a areas of monoisotopic 12C features of the respective feature pairs, b areas of U-13C labelled features, c intensity ratios of monoisotopic 12C and corresponding U-13C feature area (internal standardisation) [figure-width: 174 mm]
Fig. 5
Histograms showing the distributions of coefficients of variation (CV) across all SIL derived features which were consistently found in all replicates of F. graminearum wildtype PH-1 (n = 6) and F. graminearum aggregate samples (n = 13). The histograms in a and b (red) were derived from the peak areas of the monoisotopic 12C feature of the respective feature pairs while c and d (blue) were calculated after internal standardisation with the areas of the corresponding U-13C labelled features of the very same feature pair. Histograms e and f (overlay of transparent red and blue) combine the respective above two histograms to illustrate the shift towards lower CVs by internal standardisation, achieved for both sample types [figure-width: 129 mm]
Similar articles
- Automated LC-HRMS(/MS) approach for the annotation of fragment ions derived from stable isotope labeling-assisted untargeted metabolomics.
Neumann NK, Lehner SM, Kluger B, Bueschl C, Sedelmaier K, Lemmens M, Krska R, Schuhmacher R. Neumann NK, et al. Anal Chem. 2014 Aug 5;86(15):7320-7. doi: 10.1021/ac501358z. Epub 2014 Jul 14. Anal Chem. 2014. PMID: 24965664 Free PMC article. - Preparation of uniformly labelled 13C- and 15N-plants using customised growth chambers.
Ćeranić A, Doppler M, Büschl C, Parich A, Xu K, Koutnik A, Bürstmayr H, Lemmens M, Schuhmacher R. Ćeranić A, et al. Plant Methods. 2020 Apr 6;16:46. doi: 10.1186/s13007-020-00590-9. eCollection 2020. Plant Methods. 2020. PMID: 32280362 Free PMC article. - Stable isotopic labelling-assisted untargeted metabolic profiling reveals novel conjugates of the mycotoxin deoxynivalenol in wheat.
Kluger B, Bueschl C, Lemmens M, Berthiller F, Häubl G, Jaunecker G, Adam G, Krska R, Schuhmacher R. Kluger B, et al. Anal Bioanal Chem. 2013 Jun;405(15):5031-6. doi: 10.1007/s00216-012-6483-8. Epub 2012 Oct 20. Anal Bioanal Chem. 2013. PMID: 23086087 Free PMC article. - Isotope-coded ESI-enhancing derivatization reagents for differential analysis, quantification and profiling of metabolites in biological samples by LC/MS: A review.
Higashi T, Ogawa S. Higashi T, et al. J Pharm Biomed Anal. 2016 Oct 25;130:181-193. doi: 10.1016/j.jpba.2016.04.033. Epub 2016 Apr 30. J Pharm Biomed Anal. 2016. PMID: 27178301 Review. - From Samples to Insights into Metabolism: Uncovering Biologically Relevant Information in LC-HRMS Metabolomics Data.
Ivanisevic J, Want EJ. Ivanisevic J, et al. Metabolites. 2019 Dec 17;9(12):308. doi: 10.3390/metabo9120308. Metabolites. 2019. PMID: 31861212 Free PMC article. Review.
Cited by
- Stable Isotope-Assisted Plant Metabolomics: Investigation of Phenylalanine-Related Metabolic Response in Wheat Upon Treatment With the Fusarium Virulence Factor Deoxynivalenol.
Doppler M, Kluger B, Bueschl C, Steiner B, Buerstmayr H, Lemmens M, Krska R, Adam G, Schuhmacher R. Doppler M, et al. Front Plant Sci. 2019 Oct 30;10:1137. doi: 10.3389/fpls.2019.01137. eCollection 2019. Front Plant Sci. 2019. PMID: 31736983 Free PMC article. - Automated LC-HRMS(/MS) approach for the annotation of fragment ions derived from stable isotope labeling-assisted untargeted metabolomics.
Neumann NK, Lehner SM, Kluger B, Bueschl C, Sedelmaier K, Lemmens M, Krska R, Schuhmacher R. Neumann NK, et al. Anal Chem. 2014 Aug 5;86(15):7320-7. doi: 10.1021/ac501358z. Epub 2014 Jul 14. Anal Chem. 2014. PMID: 24965664 Free PMC article. - High-throughput Saccharomyces cerevisiae cultivation method for credentialing-based untargeted metabolomics.
Favilli L, Griffith CM, Schymanski EL, Linster CL. Favilli L, et al. Anal Bioanal Chem. 2023 Jul;415(17):3415-3434. doi: 10.1007/s00216-023-04724-5. Epub 2023 May 22. Anal Bioanal Chem. 2023. PMID: 37212869 Free PMC article. - QCScreen: a software tool for data quality control in LC-HRMS based metabolomics.
Simader AM, Kluger B, Neumann NK, Bueschl C, Lemmens M, Lirk G, Krska R, Schuhmacher R. Simader AM, et al. BMC Bioinformatics. 2015 Oct 24;16:341. doi: 10.1186/s12859-015-0783-x. BMC Bioinformatics. 2015. PMID: 26498454 Free PMC article. - Lack of the COMPASS Component Ccl1 Reduces H3K4 Trimethylation Levels and Affects Transcription of Secondary Metabolite Genes in Two Plant-Pathogenic Fusarium Species.
Studt L, Janevska S, Arndt B, Boedi S, Sulyok M, Humpf HU, Tudzynski B, Strauss J. Studt L, et al. Front Microbiol. 2017 Jan 9;7:2144. doi: 10.3389/fmicb.2016.02144. eCollection 2016. Front Microbiol. 2017. PMID: 28119673 Free PMC article.
References
- Böttcher C, Roepenack-Lahaye EV, Willscher E, Scheel D, Clemens S. Evaluation of matrix effects in metabolite profiling based on capillary liquid chromatography electrospray ionization quadrupole time-of-flight mass spectrometry. Analytical Chemistry. 2007;79:1507–1513. doi: 10.1021/ac061037q. - DOI - PubMed
- Brown M, Dunn WB, Dobson P, Patel Y, Winder CL, Francis-Mcintyre S, Begley P, Carroll K, Broadhurst D, Tseng A, Swainston N, Spasic I, Goodacre R, Kell DB. Mass spectrometry tools and metabolite-specific databases for molecular identification in metabolomics. Analyst. 2009;134:1322–1332. doi: 10.1039/b901179j. - DOI - PubMed
- Bueschl C, Kluger B, Berthiller F, Lirk G, Winkler S, Krska R, Schuhmacher R. MetExtract: A new software tool for the automated comprehensive extraction of metabolite-derived LC/MS signals in metabolomics research. Bioinformatics. 2012;28:736–738. doi: 10.1093/bioinformatics/bts012. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources