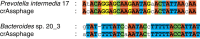A highly abundant bacteriophage discovered in the unknown sequences of human faecal metagenomes - PubMed (original) (raw)
Noriko Cassman 2, Katelyn McNair 3, Savannah E Sanchez 4, Genivaldo G Z Silva 5, Lance Boling 4, Jeremy J Barr 4, Daan R Speth 6, Victor Seguritan 4, Ramy K Aziz 7, Ben Felts 8, Elizabeth A Dinsdale 9, John L Mokili 4, Robert A Edwards 10
Affiliations
- PMID: 25058116
- PMCID: PMC4111155
- DOI: 10.1038/ncomms5498
A highly abundant bacteriophage discovered in the unknown sequences of human faecal metagenomes
Bas E Dutilh et al. Nat Commun. 2014.
Abstract
Metagenomics, or sequencing of the genetic material from a complete microbial community, is a promising tool to discover novel microbes and viruses. Viral metagenomes typically contain many unknown sequences. Here we describe the discovery of a previously unidentified bacteriophage present in the majority of published human faecal metagenomes, which we refer to as crAssphage. Its ~97 kbp genome is six times more abundant in publicly available metagenomes than all other known phages together; it comprises up to 90% and 22% of all reads in virus-like particle (VLP)-derived metagenomes and total community metagenomes, respectively; and it totals 1.68% of all human faecal metagenomic sequencing reads in the public databases. The majority of crAssphage-encoded proteins match no known sequences in the database, which is why it was not detected before. Using a new co-occurrence profiling approach, we predict a Bacteroides host for this phage, consistent with Bacteroides-related protein homologues and a unique carbohydrate-binding domain encoded in the phage genome.
Figures
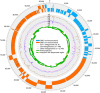
Figure 1. Schematic representation of the circular crAssphage genome.
The genome contains 80 ORFs that were predicted with Glimmer trained on_Caudovirales_. The total coverage of each nucleotide in the F2T1 metagenome, and in all public metagenomes in MG-RAST is indicated (466 human faecal and 2,440 other metagenomes, as determined by blastn mapping: ≥75 bp aligned with ≥95% identity, see Methods). Green bars indicate the 36 regions that were validated by long-range PCR (see Table 2 and Supplementary Table 1). Selected regions of several PCR amplicons (indicated as light green regions in the green bars) were sequenced by Sanger dideoxynucleotide sequencing to validate that the amplicons were indeed derived from the crAssphage genome (Supplementary Table 1). See Supplementary Fig. 6 for the fully annotated figure.
Figure 2. CRISPR spacers similar to regions of the crAssphage genome.
CRISPR spacers were identified in 2,773 complete bacterial genomes from Genbank, and in 404 genomes of intestinal isolates from HMP and MetaHIT. The CRISPR spacers that were most similar to the crAssphage genome were found in Prevotella intermedia 17 (Genbank genomes) and in_Bacteroides_ sp. 20_3 (HMP and MetaHIT genomes). Conserved A, C, G, and T nucleotides are displayed in red, green, yellow and blue, respectively.
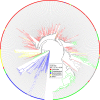
Figure 3. Phage–host prediction based on co-occurrence across metagenomes.
Unrooted co-occurrence cladogram of correlated depth profiles across 151 HMP faecal metagenomes of the crAssphage, two known _Bacteroides fragilis_-infecting phages, and 404 potential hosts. Colours indicate bacterial phyla. The phages are indicated with blue dashed lines. See Supplementary Fig. 7 for the fully annotated figure.
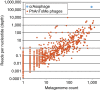
Figure 4. Abundance ubiquity plot of phage genomes in public metagenomes.
Reads from 2,944 publicly available shotgun metagenomes were aligned to a database of 1,193 phage genomes (see Methods). The average depth of aligned reads per nucleotide of the phage genome (abundance) is plotted against the number of metagenomes it is found in (ubiquity). See Supplementary Data 5 for details.
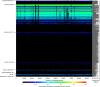
Figure 5. Normalized coverage plot of the crAssphage genome in 940 public metagenomes.
Rows are metagenomes, with the sequence volume in nucleotides indicated to the right (see Supplementary Data 4 for the order and detailed annotations of the metagenomes). The x axis of the heat map displays the 97,065 bp length of the crAssphage genome sequence. The colour bar indicates the percentage of nucleotides in each metagenome that aligns to each position. Black arrowheads at the top of the figure indicate metaviromic islands. Details are available in Supplementary Data 4. Note that some of the metagenomes at the bottom of the plot that are annotated as ‘Plant-associated’ are also faecal metagenomes.
Similar articles
- Evolution of BACON Domain Tandem Repeats in crAssphage and Novel Gut Bacteriophage Lineages.
Jonge PA, Meijenfeldt FABV, Rooijen LEV, Brouns SJJ, Dutilh BE. Jonge PA, et al. Viruses. 2019 Nov 21;11(12):1085. doi: 10.3390/v11121085. Viruses. 2019. PMID: 31766550 Free PMC article. - Microbial Diversity and Phage-Host Interactions in the Georgian Coastal Area of the Black Sea Revealed by Whole Genome Metagenomic Sequencing.
Jaiani E, Kusradze I, Kokashvili T, Geliashvili N, Janelidze N, Kotorashvili A, Kotaria N, Guchmanidze A, Tediashvili M, Prangishvili D. Jaiani E, et al. Mar Drugs. 2020 Nov 14;18(11):558. doi: 10.3390/md18110558. Mar Drugs. 2020. PMID: 33202695 Free PMC article. - Comparative analysis of CRISPR cassettes from the human gut metagenomic contigs.
Gogleva AA, Gelfand MS, Artamonova II. Gogleva AA, et al. BMC Genomics. 2014 Mar 17;15(1):202. doi: 10.1186/1471-2164-15-202. BMC Genomics. 2014. PMID: 24628983 Free PMC article. - CrAss-Like Phages: From Discovery in Human Fecal Metagenome to Application as a Microbial Source Tracking Marker.
Remesh AT, Viswanathan R. Remesh AT, et al. Food Environ Virol. 2024 Jun;16(2):121-135. doi: 10.1007/s12560-024-09584-5. Epub 2024 Feb 27. Food Environ Virol. 2024. PMID: 38413544 Review. - Computational approaches to predict bacteriophage-host relationships.
Edwards RA, McNair K, Faust K, Raes J, Dutilh BE. Edwards RA, et al. FEMS Microbiol Rev. 2016 Mar;40(2):258-72. doi: 10.1093/femsre/fuv048. Epub 2015 Dec 9. FEMS Microbiol Rev. 2016. PMID: 26657537 Free PMC article. Review.
Cited by
- Diversity and evolution of B-family DNA polymerases.
Kazlauskas D, Krupovic M, Guglielmini J, Forterre P, Venclovas Č. Kazlauskas D, et al. Nucleic Acids Res. 2020 Oct 9;48(18):10142-10156. doi: 10.1093/nar/gkaa760. Nucleic Acids Res. 2020. PMID: 32976577 Free PMC article. Review. - Assembly of viral genomes from metagenomes.
Smits SL, Bodewes R, Ruiz-Gonzalez A, Baumgärtner W, Koopmans MP, Osterhaus AD, Schürch AC. Smits SL, et al. Front Microbiol. 2014 Dec 18;5:714. doi: 10.3389/fmicb.2014.00714. eCollection 2014. Front Microbiol. 2014. PMID: 25566226 Free PMC article. - Environmental genes and genomes: understanding the differences and challenges in the approaches and software for their analyses.
Zepeda Mendoza ML, Sicheritz-Pontén T, Gilbert MT. Zepeda Mendoza ML, et al. Brief Bioinform. 2015 Sep;16(5):745-58. doi: 10.1093/bib/bbv001. Epub 2015 Feb 11. Brief Bioinform. 2015. PMID: 25673291 Free PMC article. - The dark side of the gut: Virome-host interactions in intestinal homeostasis and disease.
Li Y, Handley SA, Baldridge MT. Li Y, et al. J Exp Med. 2021 May 3;218(5):e20201044. doi: 10.1084/jem.20201044. J Exp Med. 2021. PMID: 33760921 Free PMC article. Review. - Functional characterization of prokaryotic dark matter: the road so far and what lies ahead.
Escudeiro P, Henry CS, Dias RPM. Escudeiro P, et al. Curr Res Microb Sci. 2022 Aug 7;3:100159. doi: 10.1016/j.crmicr.2022.100159. eCollection 2022. Curr Res Microb Sci. 2022. PMID: 36561390 Free PMC article. Review.
References
- Tyson G. W. et al.. Community structure and metabolism through reconstruction of microbial genomes from the environment. Nature 428, 37–43 (2004). - PubMed
- Cassman N. et al.. Oxygen minimum zones harbour novel viral communities with low diversity. Environ. Microbiol. 14, 3043–3065 (2012). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical