Requirement for PBAF in transcriptional repression and repair at DNA breaks in actively transcribed regions of chromatin - PubMed (original) (raw)
Requirement for PBAF in transcriptional repression and repair at DNA breaks in actively transcribed regions of chromatin
Andreas Kakarougkas et al. Mol Cell. 2014.
Abstract
Actively transcribed regions of the genome are vulnerable to genomic instability. Recently, it was discovered that transcription is repressed in response to neighboring DNA double-strand breaks (DSBs). It is not known whether a failure to silence transcription flanking DSBs has any impact on DNA repair efficiency or whether chromatin remodelers contribute to the process. Here, we show that the PBAF remodeling complex is important for DSB-induced transcriptional silencing and promotes repair of a subset of DNA DSBs at early time points, which can be rescued by inhibiting transcription globally. An ATM phosphorylation site on BAF180, a PBAF subunit, is required for both processes. Furthermore, we find that subunits of the PRC1 and PRC2 polycomb group complexes are similarly required for DSB-induced silencing and promoting repair. Cancer-associated BAF180 mutants are unable to restore these functions, suggesting PBAF's role in repressing transcription near DSBs may contribute to its tumor suppressor activity.
Copyright © 2014 The Authors. Published by Elsevier Inc. All rights reserved.
Figures
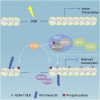
Graphical abstract
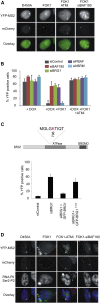
Figure 1
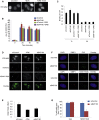
PBAF Is Required for Transcriptional Silencing Induced by DNA DSBs (A) Representative immunofluorescence images of U2OS reporter cells expressing mCherry-tagged WT or nuclease-deficient (D450A) FokI (Shanbhag et al., 2010). On addition of doxycycline, YFP signal accumulation at the reporter site is a result of YFP-MS2 protein binding to MS2 stem loops and is indicative of nascent transcript formation. Following transfection of WT FokI, but not the nuclease-deficient Fok1 (D450A), DSBs are induced at the reporter site and transcriptional silencing occurs, leading to loss of YFP signal. Treatment with ATMi or siBAF180 leads to persistent YFP signal formation at the reporter site in WT FokI-expressing cells indicating deficient transcriptional silencing in response to DNA DSBs. (B) Quantification of doxycycline (DOX)-induced transcription in U2OS reporter cells treated with the indicated siRNA. Transcriptional silencing was monitored after addition of doxycycline by quantification of YFP-positive cells expressing WT FokI endonuclease (FOK1) in the presence and absence of ATMi. Data are represented as mean ± SD. (C) Top: cartoon showing the location of K798 in the catalytic subunit of PBAF (BRG1). Bottom: siRNA-resistant WT or K798R mutant BRG1 expression constructs were introduced into siBRG1 cells and assayed for transcriptional silencing in cells expressing WT FokI endonuclease by quantification of YFP-positive cells. Data are represented as mean ± SD. BROMO, bromodomain. (D) Actively elongating RNA polymerase II (RNAPII) at the reporter site was monitored using an antibody against phosphorylated Ser2 of the C-terminal domain of the RNAPII large subunit. See also Figure S1.
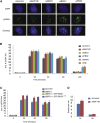
Figure 2
PBAF Contributes to NHEJ at Early Time Points following DNA Damage (A) Representative immunofluorescence images of A549 cells treated with the indicated siRNA, 40 min following exposure to 1.5 Gy IR. (B) Quantification of γH2AX foci clearance following exposure to 1.5 Gy IR in A549 cells treated with the indicated siRNA or ATMi. Data are represented as mean ± SD. h, hours; Ave., average. (C) siRNA-resistant WT or K798R mutant BRG1 expression constructs were introduced into siBRG1 cells and assayed for γH2AX foci clearance following exposure to 1.5 Gy IR. Data are represented as mean ± SD. (D) Chromosome breakage analysis in siControl- and siBAF180-treated 82-6 hTert fibroblasts at early and late times following 7 Gy IR. Data are represented as mean ± SE. See also Figures S2 and S3.
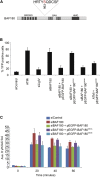
Figure 3
Phosphorylation of BAF180 by ATM Is Required for Transcriptional Repression and Early DNA Repair Activity (A) Cartoon of human BAF180 highlighting the domain organization and the sequence surrounding serine 963 (Ser 948 in isoform 1), which is phosphorylated by ATM in response to DNA damage. (B) Quantification of YFP-positive cells as a measure of DSB-induced transcriptional repression in U2OS reporter cells treated with the indicated siRNA and complemented with BAF180 expression constructs as indicated. Data are represented as mean ± SD. (C) Quantification of γH2AX foci clearance following exposure to 1.5 Gy IR in U2OS cells treated with the indicated siRNA and complemented with BAF180 expression constructs as indicated. Data are represented as mean ± SD. Ave., average. See also Figures S1 and S4.
Figure 4
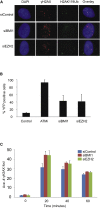
PBAF Is Required for Efficient Repair Only when There Is Ongoing Transcription and Promotes H2A K119 Ubiquitination at DSBs (A) Treatment of U2OS reporter cells with DRB efficiently inhibits transcription, as demonstrated by the loss of YFP signal at the reporter site of doxycycline-treated cells. (B) Quantification of γH2AX foci clearance following exposure to 1.5 Gy IR in control and siBAF180 A549 cells with and without DRB treatment prior to IR. Data are represented as mean ± SD. Ave., average. (C) Chromosome breakage analysis in siControl- and siBAF180-treated 82-6 hTert fibroblasts at early and late times following 7 Gy IR with the addition of DRB (data without DRB are as in Figure 2C). Data are represented as mean ± SE. h, hours. (D) Representative immunofluorescence images of YFP, mCherry, and H2AK119ub following DSB induction in control, ATM inhibitor-treated, or siBAF180 cells. (E) Relative mean fluorescence intensity (RMFI) of H2AK119Ub signal at the reporter site of WT FokI expressing cells treated with ATMi or siBAF180, relative to control cells. Data are represented as mean ± SD. (F) The formation of conjugated ubiquitin IR-induced foci (IRIF) (FK2) and 53BP1 IRIF is unaffected by siBAF180 in A549 cells exposed to 1.5 Gy IR and fixed 1 hr post-IR. (G) Quantification of FK2 and H2A K119Ub IRIF in A549 cells exposed to 1.5 Gy IR and fixed 1 hr post-IR. Data are represented as mean ± SD. See also Figure S5.
Figure 5
BMI1 and EZH2, Subunits of PRC1 and PRC2, Respectively, Are Required for DSB-Induced H2AK119ub and Transcriptional Silencing and Efficient DSB Repair at Early Time Points (A) The formation of H2AK119ub IR-induced foci (IRIF) is defective in cells depleted for either BMI1 or EZH2 in A549 cells, 60 min following exposure to 1.5 Gy IR. (B) Quantification of YFP-positive cells as a measure of DSB-induced transcriptional repression in U2OS reporter cells treated with the indicated siRNA. Data are represented as mean ± SD. (C) Quantification of γH2AX foci clearance following exposure to 1.5 Gy IR in U2OS cells treated with the indicated siRNA. Data are represented as mean ± SD. Ave., average. See also Figure S6.
Figure 6
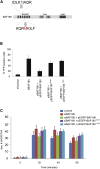
Cancer-Associated Mutations of BAF180 Do Not Complement DSB Repair and Transcriptional Silencing Defects Associated with Loss of BAF180 (A) Schematic representation of BAF180, showing the positions of the T232P and M538I mutations identified in ccRCC samples. (B) Quantification of YFP-positive cells as a measure of DSB-induced transcriptional silencing in U2OS reporter cells treated with BAF180 or control siRNA and complemented with BAF180 expression constructs as indicated. Data are represented as mean ± SD. (C) Quantification of γH2AX foci clearance following exposure to 1.5 Gy IR in U2OS cells treated with BAF180 or control siRNA and complemented with BAF180 expression constructs as indicated. Data are represented as mean ± SD. See also Figure S1.
Similar articles
- The chromatin-remodeling subunit Baf200 promotes homology-directed DNA repair and regulates distinct chromatin-remodeling complexes.
de Castro RO, Previato L, Goitea V, Felberg A, Guiraldelli MF, Filiberti A, Pezza RJ. de Castro RO, et al. J Biol Chem. 2017 May 19;292(20):8459-8471. doi: 10.1074/jbc.M117.778183. Epub 2017 Apr 5. J Biol Chem. 2017. PMID: 28381560 Free PMC article. - Remodeling and spacing factor 1 (RSF1) deposits centromere proteins at DNA double-strand breaks to promote non-homologous end-joining.
Helfricht A, Wiegant WW, Thijssen PE, Vertegaal AC, Luijsterburg MS, van Attikum H. Helfricht A, et al. Cell Cycle. 2013 Sep 15;12(18):3070-82. doi: 10.4161/cc.26033. Epub 2013 Aug 20. Cell Cycle. 2013. PMID: 23974106 Free PMC article. - The SWI/SNF ATPase BRG1 stimulates DNA end resection and homologous recombination by reducing nucleosome density at DNA double strand breaks and by promoting the recruitment of the CtIP nuclease.
Hays E, Nettleton E, Carter C, Morales M, Vo L, Passo M, Vélez-Cruz R. Hays E, et al. Cell Cycle. 2020 Nov;19(22):3096-3114. doi: 10.1080/15384101.2020.1831256. Epub 2020 Oct 12. Cell Cycle. 2020. PMID: 33044911 Free PMC article. - Chromatin modification and NBS1: their relationship in DNA double-strand break repair.
Saito Y, Zhou H, Kobayashi J. Saito Y, et al. Genes Genet Syst. 2016;90(4):195-208. doi: 10.1266/ggs.15-00010. Epub 2015 Nov 25. Genes Genet Syst. 2016. PMID: 26616756 Review. - The influence of heterochromatin on DNA double strand break repair: Getting the strong, silent type to relax.
Goodarzi AA, Jeggo P, Lobrich M. Goodarzi AA, et al. DNA Repair (Amst). 2010 Dec 10;9(12):1273-82. doi: 10.1016/j.dnarep.2010.09.013. Epub 2010 Oct 30. DNA Repair (Amst). 2010. PMID: 21036673 Review.
Cited by
- Prognostic and Predictive Value of PBRM1 in Clear Cell Renal Cell Carcinoma.
Carril-Ajuria L, Santos M, Roldán-Romero JM, Rodriguez-Antona C, de Velasco G. Carril-Ajuria L, et al. Cancers (Basel). 2019 Dec 19;12(1):16. doi: 10.3390/cancers12010016. Cancers (Basel). 2019. PMID: 31861590 Free PMC article. Review. - BRG1 Promotes chromatin remodeling around DNA damage sites.
Qi W, Chen H, Lu C, Bu Q, Wang X, Han L. Qi W, et al. Anim Cells Syst (Seoul). 2018 Oct 1;22(6):360-367. doi: 10.1080/19768354.2018.1525429. eCollection 2018. Anim Cells Syst (Seoul). 2018. PMID: 30533258 Free PMC article. - Loss of PBRM1 Alters Promoter Histone Modifications and Activates ALDH1A1 to Drive Renal Cell Carcinoma.
Schoenfeld DA, Zhou R, Zairis S, Su W, Steinbach N, Mathur D, Bansal A, Zachem AL, Tavarez B, Hasson D, Bernstein E, Rabadan R, Parsons R. Schoenfeld DA, et al. Mol Cancer Res. 2022 Aug 5;20(8):1193-1207. doi: 10.1158/1541-7786.MCR-21-1039. Mol Cancer Res. 2022. PMID: 35412614 Free PMC article. - Mammalian SWI/SNF collaborates with a polycomb-associated protein to regulate male germline transcription in the mouse.
Menon DU, Shibata Y, Mu W, Magnuson T. Menon DU, et al. Development. 2019 Jul 5;146(19):dev174094. doi: 10.1242/dev.174094. Development. 2019. PMID: 31043422 Free PMC article. - Hypoxia and Chromatin: A Focus on Transcriptional Repression Mechanisms.
Batie M, Del Peso L, Rocha S. Batie M, et al. Biomedicines. 2018 Apr 22;6(2):47. doi: 10.3390/biomedicines6020047. Biomedicines. 2018. PMID: 29690561 Free PMC article. Review.
References
- Cao R., Tsukada Y., Zhang Y. Role of Bmi-1 and Ring1A in H2A ubiquitylation and Hox gene silencing. Mol. Cell. 2005;20:845–854. - PubMed
- Chambers A.L., Downs J.A. The RSC and INO80 chromatin-remodeling complexes in DNA double-strand break repair. Prog. Mol. Biol. Transl. Sci. 2012;110:229–261. - PubMed
- Chou D.M., Adamson B., Dephoure N.E., Tan X., Nottke A.C., Hurov K.E., Gygi S.P., Colaiácovo M.P., Elledge S.J. A chromatin localization screen reveals poly (ADP ribose)-regulated recruitment of the repressive polycomb and NuRD complexes to sites of DNA damage. Proc. Natl. Acad. Sci. USA. 2010;107:18475–18480. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous