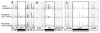Identification of host genes that affect acquisition of an integrative and conjugative element in Bacillus subtilis - PubMed (original) (raw)
. 2014 Sep;93(6):1284-301.
doi: 10.1111/mmi.12736. Epub 2014 Aug 15.
Affiliations
- PMID: 25069588
- PMCID: PMC4160349
- DOI: 10.1111/mmi.12736
Identification of host genes that affect acquisition of an integrative and conjugative element in Bacillus subtilis
Christopher M Johnson et al. Mol Microbiol. 2014 Sep.
Erratum in
- Identification of host genes that affect acquisition of an integrative and conjugative element in Bacillus subtilis.
Johnson CM, Grossman AD. Johnson CM, et al. Mol Microbiol. 2015 Dec;98(6):1222. doi: 10.1111/mmi.13282. Mol Microbiol. 2015. PMID: 26767850 No abstract available.
Abstract
Conjugation, a major type of horizontal gene transfer in bacteria, involves transfer of DNA from a donor to a recipient using donor-encoded conjugation machinery. Using a high-throughput screen (Tn-seq), we identified genes in recipients that contribute to acquisition of the integrative and conjugative element ICEBs1 by Bacillus subtilis. We found that null mutations in some genes caused an increase, and others a decrease in conjugation efficiency. Some mutations affected conjugation only when present in recipients. Other mutations affected conjugation when present in donors or recipients. Most of the genes identified are known or predicted to affect the cell envelope. Several encode enzymes involved in phospholipid biosynthesis and one encodes a homologue of penicillin-binding proteins. Two of the genes identified also affected conjugation of Tn916, indicating that their roles in conjugation may be general. We did not identify any genes in recipients that were essential for ICEBs1 conjugation, indicating that if there are such genes, then these are either essential for cell growth or redundant. Our results indicate that acquisition of ICEBs1, and perhaps other conjugative elements, is robust and not easily avoided by mutation and that several membrane-related functions affect the efficiency of conjugation.
© 2014 John Wiley & Sons Ltd.
Figures
Figure 1
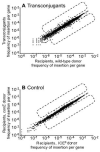
Tn-seq analysis to identify genes in recipients that affect acquisition of ICE_Bs1_. The frequencies of transposon insertions in annotated genes in B. subtilis in different pools from the genetic screen are compared. Each dot represents a gene. The numbers on each axis represent the frequency of transposon insertions in a given gene in the indicated pool as a fraction of the total insertions sequenced from that pool. Dashed boxes are used to visually approximate the criteria used to identify candidate genes. A. The frequency of transposon insertions in each gene in the transconjugant population (Y axis) plotted versus the recipient population from the same mating, irrespective of ICE_Bs1_ acquisition (X axis). B. The frequency of transposon insertions in each gene in the recipient population from a mock-mating with the conjugation-defective donor MMB963 (Y axis) plotted versus the recipient population from a mock-mating with the ICE_Bs1_0 donor JMA222 (X axis).
Figure 2
Locations of transposon insertions in representative candidate genes. Each histogram shows the location and frequency of transposon insertions in the indicated genome region. A. mprF; B. yfnI; C. yonF. The top row shows insertions in the transconjugant, the middle and bottom rows show insertions in the indicated control pools. Middle: Recipients from mating with WT (wild type) donor (KM250); bottom: recipients from a mock-mating with ICE_Bs1_0 donor (JMA222). The location on the X axis represents the location on the chromosome, and the height of each line represents the number of transposon insertions at that location, relative to the number of insertions at other locations in the region. The boxed regions denote the boundaries of genes of interest. The map on the bottom shows the location and orientation of genes. The gene of interest is filled in black.
Figure 3
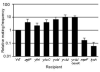
Effects of mutations in the recipient on acquisition of ICE_Bs1_. Standard filter matings were performed using a wild type donor (KM250). Recipients were WT (CMJ161), or had null mutations in ugtP (CMJ83); yfnI (CMJ44); ykuC (CMJ46); yvbI (CMJ309); yvbJ (CMJ153); yvbJ and comK (CMJ179); mprF (CMJ162); lysA (CMJ335, grown with 40 μg/ml lysine). The relative mating frequency (y-axis) is the number of transconjugants per donor of the indicated strain normalized to that of wild type. The wild type mating efficiency was 0.048 (4.8°) transconjugants per donor ± 0.053 (standard deviation). Graphs show means and standard deviation from ≥3 experiments. For all mutants, p ≤ 0.05, _t_-test, indicating that the conjugation frequency of each mutant is likely to be different from that of wild type.
Figure 4
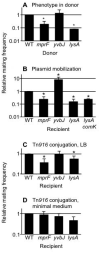
Effects of host mutations on conjugation and plasmid mobilization. Mutants were tested in standard filter matings for their phenotype on the ability of a donor to deliver ICE_Bs1_. Relative mating frequencies are presented as a fraction of that of wild type donor and wild type recipient, as described for Fig. 3. Data presented are the averages ± standard deviation from ≥3 experiments. Asterisks above bars indicate a p value ≤0.05 in a t-test. A. The indicated mutations were present in donor and ICE_Bs1 was transferred to wild type recipients (CAL89). Donors strains were WT (KM250), or had null mutations in mprF (CMJ323), yvbJ (CMJ186), or lysA (CMJ351, grown with 40 μg/ml lysine). The average and standard deviation of mating with the wild type donor was 0.019 ± 0.036 transconjugants per donor. B. Mobilization of pC194 by ICE_Bs1 into mutant recipients. A strain (CMJ300) containing ICE_Bs1_ and pC194 was used as donor. Recipients were WT (CMJ161) or had null mutations in mprF (CMJ162), yvbJ (CMJ153), lysA (CMJ335) or lysA and comK (CMJ386). lysA mutants were grown in medium supplemented with 40 μg/ml lysine. Average and standard deviation of mobilization of pC194 into the WT recipient was 0.0045 ± 0.0030. C, D. Transfer of Tn_916_ into mutant recipients. Donors containing Tn_916_ (CM253) and indicated recipients were grown in (C) LB medium or (D) minimal medium prior to mating. Recipients were WT (CAL89) or had null mutations in mprF (CMJ162), yvbJ (CMJ153) or lysA (CMJ335, grown with 40 μg/ml lysine). Average and standard deviation of transfer of Tn_916_ into the WT recipient was 0.00025 ± 0.00010 for cells grown in LB and 0.00016 ± 0.00006 for cells grown in minimal medium.
Figure 5
YvbJ is paralogous to PBP4a. A global alignment between YvbJ and PBP4a is shown. Features shown include: the transmembrane domain (TM) predicted by TMHMM and sequence signatures of the penicilloyl serine transferases superfamily: 1: SxxK motif; 2: (S/Y)xN motif; 3: K(T/S)G motif.
Similar articles
- Characterization of ConE, the VirB4 Homolog of the Integrative and Conjugative Element ICE_Bs1_ of Bacillus subtilis.
Murthy AC, Aleksanyan N, Morton GM, Toyoda HC, Kalashyan M, Chen S, Ragucci AE, Broulidakis MP, Swerdlow KJ, Bui MNN, Muccioli M, Berkmen MB. Murthy AC, et al. J Bacteriol. 2023 Jun 27;205(6):e0003323. doi: 10.1128/jb.00033-23. Epub 2023 May 23. J Bacteriol. 2023. PMID: 37219457 Free PMC article. - Specificity and Selective Advantage of an Exclusion System in the Integrative and Conjugative Element ICE_Bs1_ of Bacillus subtilis.
Davis KP, Grossman AD. Davis KP, et al. J Bacteriol. 2021 Apr 21;203(10):e00700-20. doi: 10.1128/JB.00700-20. Print 2021 Apr 21. J Bacteriol. 2021. PMID: 33649151 Free PMC article. - A CRISPR interference screen reveals a role for cell wall teichoic acids in conjugation in Bacillus subtilis.
Harden MM, Anderson ME, Grossman AD. Harden MM, et al. Mol Microbiol. 2022 Jun;117(6):1366-1383. doi: 10.1111/mmi.14914. Epub 2022 May 21. Mol Microbiol. 2022. PMID: 35490406 Free PMC article. - Biology of ICEBs1, an integrative and conjugative element in Bacillus subtilis.
Auchtung JM, Aleksanyan N, Bulku A, Berkmen MB. Auchtung JM, et al. Plasmid. 2016 Jul;86:14-25. doi: 10.1016/j.plasmid.2016.07.001. Epub 2016 Jul 2. Plasmid. 2016. PMID: 27381852 Review. - Integrative and Conjugative Elements (ICEs): What They Do and How They Work.
Johnson CM, Grossman AD. Johnson CM, et al. Annu Rev Genet. 2015;49:577-601. doi: 10.1146/annurev-genet-112414-055018. Epub 2015 Oct 14. Annu Rev Genet. 2015. PMID: 26473380 Free PMC article. Review.
Cited by
- TnSmu1 is a functional integrative and conjugative element in Streptococcus mutans that when expressed causes growth arrest of host bacteria.
McLellan LK, Anderson ME, Grossman AD. McLellan LK, et al. Mol Microbiol. 2022 Dec;118(6):652-669. doi: 10.1111/mmi.14992. Epub 2022 Nov 11. Mol Microbiol. 2022. PMID: 36268794 Free PMC article. - Implementation and Data Analysis of Tn-seq, Whole-Genome Resequencing, and Single-Molecule Real-Time Sequencing for Bacterial Genetics.
Burby PE, Nye TM, Schroeder JW, Simmons LA. Burby PE, et al. J Bacteriol. 2016 Dec 13;199(1):e00560-16. doi: 10.1128/JB.00560-16. Print 2017 Jan 1. J Bacteriol. 2016. PMID: 27672193 Free PMC article. Review. - Identification of Genes Required for Swarming Motility in Bacillus subtilis Using Transposon Mutagenesis and High-Throughput Sequencing (TnSeq).
Sanchez S, Snider EV, Wang X, Kearns DB. Sanchez S, et al. J Bacteriol. 2022 Jun 21;204(6):e0008922. doi: 10.1128/jb.00089-22. Epub 2022 May 31. J Bacteriol. 2022. PMID: 35638827 Free PMC article. - Discovery of a dual protease mechanism that promotes DNA damage checkpoint recovery.
Burby PE, Simmons ZW, Schroeder JW, Simmons LA. Burby PE, et al. PLoS Genet. 2018 Jul 6;14(7):e1007512. doi: 10.1371/journal.pgen.1007512. eCollection 2018 Jul. PLoS Genet. 2018. PMID: 29979679 Free PMC article. - Critical Components of the Conjugation Machinery of the Integrative and Conjugative Element ICEBs1 of Bacillus subtilis.
Leonetti CT, Hamada MA, Laurer SJ, Broulidakis MP, Swerdlow KJ, Lee CA, Grossman AD, Berkmen MB. Leonetti CT, et al. J Bacteriol. 2015 Aug 1;197(15):2558-67. doi: 10.1128/JB.00142-15. Epub 2015 May 26. J Bacteriol. 2015. PMID: 26013486 Free PMC article.
References
- Akerley BJ, Lampe DJ. Analysis of gene function in bacterial pathogens by GAMBIT. Methods in Enzymology. 2002;358:100–108. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases