Molecular architecture and function of the SEA complex, a modulator of the TORC1 pathway - PubMed (original) (raw)
Molecular architecture and function of the SEA complex, a modulator of the TORC1 pathway
Romain Algret et al. Mol Cell Proteomics. 2014 Nov.
Abstract
The TORC1 signaling pathway plays a major role in the control of cell growth and response to stress. Here we demonstrate that the SEA complex physically interacts with TORC1 and is an important regulator of its activity. During nitrogen starvation, deletions of SEA complex components lead to Tor1 kinase delocalization, defects in autophagy, and vacuolar fragmentation. TORC1 inactivation, via nitrogen deprivation or rapamycin treatment, changes cellular levels of SEA complex members. We used affinity purification and chemical cross-linking to generate the data for an integrative structure modeling approach, which produced a well-defined molecular architecture of the SEA complex and showed that the SEA complex comprises two regions that are structurally and functionally distinct. The SEA complex emerges as a platform that can coordinate both structural and enzymatic activities necessary for the effective functioning of the TORC1 pathway.
© 2014 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures
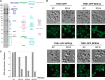
Fig. 1.
Identification of SEA complex interconnectivity and domain interaction by immunoprecipitation and chemical cross-linking. A, immunoprecipitation of Protein A (PrA) tagged proteins (indicated and underlined on the top of the gel lanes) was performed as described under “Experimental Procedures.” SEA complex proteins and their partners were resolved via SDS-PAGE and visualized with Coomassie Blue staining. Proteins identified via mass spectrometry are marked by solid circles at the right of the gel lane and are listed in order below (S1 = Sea1, S2 = Sea2, S3 = Sea3, S4 = Sea4, N3 = Npr3). PrA tagged proteins are indicated in red, co-purifying proteins in black, and IgG contaminant in gray. Shown are only members of the SEA complex. For the complete set of co-purifying proteins in lanes 6, 8, 12, 18, and 20, see
supplemental Fig. S2
and
supplemental Table S3
. The identity of a truncated protein (in amino acid residues) or deleted SEA member is indicated on the top of the gel lane. WT, wild type; Sea1G (lane 2) is a fraction from the sucrose gradient gel (
supplemental Fig. S1
). Each individual gel image was differentially scaled along its length so that its molecular mass standards aligned to a single reference set of molecular mass standards. Contrast was adjusted to improve visibility. All original gel figures are available upon request. B, co-purification profile of different SEA deletion and truncation strains. Horizontal gray lines represent the number of amino acid residues in each protein; amino acid residue positions are shown on top of the lines. Co-purifying SEA complex proteins are indicated by “+,” and missing proteins by “−.” The Sea1, Npr2, and Npr3 proteins are colored in blue; others are in yellow. C, summary of identified interprotein cross-links of the SEA complex, generated using AUTOCAD (Autodesk INC., educational version). A representative high-resolution MS/MS spectrum of a cross-linked peptide connecting two different proteins (inter-cross-link) of the SEA complex is shown on the right. An example MS spectra is shown in which the cross-linking site Sea3(1072)-Sea4(885) is unambiguously identified.
Fig. 2.
The four-stage scheme for integrative structure determination of the SEA complex. Our integrative approach proceeds through four stages (, –31): (i) gathering of data, (ii) representation of subunits and translation of data into spatial restraints, (iii) configurational sampling to produce an ensemble of models that satisfy the restraints, and (iv) analysis and assessment of the ensemble. The modeling protocol (i.e. Stages 2, 3, and 4) was scripted using the Python Modeling Interface, version 47dafcc, a library to model macromolecular complexes based on our open source Integrative Modeling Platform package, version 65734ec (32).
Fig. 3.
Molecular architecture and contact frequency of the SEA complex. A, the molecular architecture of the SEA complex was obtained through integrative modeling based on various biochemical data (Fig. 2). The localization of each SEA complex protein is defined by a density map, contoured here at the threshold that results in 1.5 times its volume estimated from sequence (
supplemental Table S6
). Three copies of Seh1 and Sea4 were included in the complex, based on the stoichiometry data, with a symmetry constraint applied to increase the model ensemble precision. Npr2 was localized with relatively low precision, indicated by a mesh. The approximate dimensions of the SEA complex are 200 Å × 200 Å × 275 Å. B, the proximities of any two residues in the molecular architecture were measured by their relative contact frequency. A contact between a pair of residues was defined as an instance when their corresponding bead surfaces were less than 30 Å from each other. Cross-links are plotted as red dots, and the residue contact frequency is indicated by a color ranging from white (0) to dark blue (1). Each box contains the contact frequency between the corresponding pair of SEA complex proteins. C–F, Although the x-ray structures of Seh1 and Sec13 (C) and comparative models of Sea2 and Sea3 (D), Sea4 and Npr3 (E), and Sea1 and Npr2 (F) are placed inside the density map, their orientations are arbitrary; for contrast, other SEA complex proteins are shown as faint meshes.
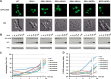
Fig. 4.
SEA complex is involved in the regulation of the TORC1 pathway. A, the yeast SEA complex physically interacts with the TORC1 complex. Sea2-PrA was immunoprecipitated as described under “Experimental Procedures.” Co-precipitating proteins were resolved via SDS-PAGE, visualized with Coomassie Blue stain, and identified via mass spectrometry (
supplemental Tables S2 and S3
). SEA complex members are marked in blue, TORC1 members in orange, mitochondria proteins in green, proteins involved in ribosome biogenesis and translation in blue, contaminants in gray, and others in pink. B, deletions of Sea1, Npr2, and Npr3 provoked Tor1 relocalization to the cytoplasm during nitrogen starvation. The localization of Tor1-GFP was followed by light fluorescence microscopy in wild-type and deletion strains of indicated SEA complex members, either in synthetic complete (SC) medium or in synthetic media lacking nitrogen SD-N, (nitrogen starvation). Observations were made for the strains grown in YPD or subjected to nitrogen starvation. C, the “vacuole-to-cytoplasm” GFP signal ratio was calculated for 25 cells in each strain shown in B.
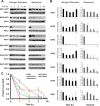
Fig. 5.
Sea1 is involved in the regulation of general autophagy. A, wild-type and indicated deletion strains transformed with a plasmid expressing GFP-ATG8 were subjected to nitrogen starvation as described under “Experimental Procedures” and examined under a fluorescent microscope after 20 h of starvation. Scale bar = 5 μm. B, strains were grown as in A. Samples were taken at the indicated time points and analyzed via Western blotting with anti-GFP or anti-PGK1 antibodies. C, autophagic flux was calculated as a ratio (percentage) of free GFP to total GFP signal (combined free GFP and GFP-ATG8) in the corresponding Western blots from B. D, to estimate autophagic induction, the total GFP signal was normalized to the PGK1 signal. Normalized values at time point 0 were set as 1.
Fig. 6.
Stability of SEA complex components during nitrogen starvation and rapamycin treatment. A, yeast cells carrying a SEA member tagged with GFP were subjected to nitrogen starvation or rapamycin treatment (20 n
m
final concentration). Samples were collected at indicated time points, and whole cell extracts were prepared and analyzed via Western blotting with anti-GFP or anti-PGK1 antibodies. Error bars represent the standard deviation in three independent experiments. B, the protein level in arbitrary units (AU) was calculated by normalizing the GFP signal to the corresponding PGK1 signal from blots shown in A. The signal at time 0 was set at 1. C, the protein level of Npr3-GFP in indicated deletion strains subjected to rapamycin treatment at different time points was calculated as in B and is represented as a graph. Error bars represent the standard deviation in three independent experiments.
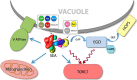
Fig. 7.
An overview of the proposed SEA complex activities and interactions. The SEA complex is situated at the vacuole membrane and interacts with V-ATPase, mitochondria, and TORC1 (straight blue arrows). The SEA complex possesses GAP activity (curved blue arrow) toward another TORC1 regulator, the EGO complex. Both SEA and EGO act upstream of the TORC1 (curved red arrows).
Similar articles
- SEA you later alli-GATOR--a dynamic regulator of the TORC1 stress response pathway.
Dokudovskaya S, Rout MP. Dokudovskaya S, et al. J Cell Sci. 2015 Jun 15;128(12):2219-28. doi: 10.1242/jcs.168922. Epub 2015 May 1. J Cell Sci. 2015. PMID: 25934700 Free PMC article. Review. - The SEACIT complex is involved in the maintenance of vacuole-mitochondria contact sites and controls mitophagy.
Ma Y, Moors A, Camougrand N, Dokudovskaya S. Ma Y, et al. Cell Mol Life Sci. 2019 Apr;76(8):1623-1640. doi: 10.1007/s00018-019-03015-6. Epub 2019 Jan 23. Cell Mol Life Sci. 2019. PMID: 30673821 Free PMC article. - The yeast chromatin remodeler Rsc1-RSC complex is required for transcriptional activation of autophagy-related genes and inhibition of the TORC1 pathway in response to nitrogen starvation.
Yu F, Imamura Y, Ueno M, Suzuki SW, Ohsumi Y, Yukawa M, Tsuchiya E. Yu F, et al. Biochem Biophys Res Commun. 2015 Sep 4;464(4):1248-1253. doi: 10.1016/j.bbrc.2015.07.114. Epub 2015 Jul 26. Biochem Biophys Res Commun. 2015. PMID: 26212438 - Conserved and Divergent Mechanisms That Control TORC1 in Yeasts and Mammals.
Morozumi Y, Shiozaki K. Morozumi Y, et al. Genes (Basel). 2021 Jan 12;12(1):88. doi: 10.3390/genes12010088. Genes (Basel). 2021. PMID: 33445779 Free PMC article. Review.
Cited by
- From integrative structural biology to cell biology.
Sali A. Sali A. J Biol Chem. 2021 Jan-Jun;296:100743. doi: 10.1016/j.jbc.2021.100743. Epub 2021 May 4. J Biol Chem. 2021. PMID: 33957123 Free PMC article. Review. - A New Crosslinking Assay to Study Guanine Nucleotide Binding in the Gtr Heterodimer of S. cerevisiae.
Doxsey DD, Veinotte K, Shen K. Doxsey DD, et al. Small GTPases. 2022 Jan;13(1):327-334. doi: 10.1080/21541248.2022.2141019. Small GTPases. 2022. PMID: 36328771 Free PMC article. - SEA you later alli-GATOR--a dynamic regulator of the TORC1 stress response pathway.
Dokudovskaya S, Rout MP. Dokudovskaya S, et al. J Cell Sci. 2015 Jun 15;128(12):2219-28. doi: 10.1242/jcs.168922. Epub 2015 May 1. J Cell Sci. 2015. PMID: 25934700 Free PMC article. Review. - The Evolving Contribution of Mass Spectrometry to Integrative Structural Biology.
Faini M, Stengel F, Aebersold R. Faini M, et al. J Am Soc Mass Spectrom. 2016 Jun;27(6):966-74. doi: 10.1007/s13361-016-1382-4. Epub 2016 Apr 7. J Am Soc Mass Spectrom. 2016. PMID: 27056566 Free PMC article. Review. - Sestrins inhibit mTORC1 kinase activation through the GATOR complex.
Parmigiani A, Nourbakhsh A, Ding B, Wang W, Kim YC, Akopiants K, Guan KL, Karin M, Budanov AV. Parmigiani A, et al. Cell Rep. 2014 Nov 20;9(4):1281-91. doi: 10.1016/j.celrep.2014.10.019. Cell Rep. 2014. PMID: 25457612 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
- T32 GM067547/GM/NIGMS NIH HHS/United States
- R01 GM083960/GM/NIGMS NIH HHS/United States
- U54 GM103511/GM/NIGMS NIH HHS/United States
- U54 GM094662/GM/NIGMS NIH HHS/United States
- P41 GM109824/GM/NIGMS NIH HHS/United States
- HHMI/Howard Hughes Medical Institute/United States
- P41 GM103314/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases