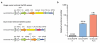Improved vectors and genome-wide libraries for CRISPR screening - PubMed (original) (raw)
Improved vectors and genome-wide libraries for CRISPR screening
Neville E Sanjana et al. Nat Methods. 2014 Aug.
No abstract available
Figures
Figure 1
New lentiviral CRISPR designs produce viruses with higher functional titer. (a) Lentiviral expression vector for Streptococcus pyogenes Cas9 and sgRNA in the improved one vector system (lentiCRISPR v2) and the two vector system (lentiCas9-Blast, lentiGuide-Puro). Psi packaging signal (psi+), rev response element (RRE), central polypurine tract (cPPT), elongation factor-1α short promoter (EFS), FLAG octapeptide tag (FLAG), 2A self-cleaving peptide (P2A), puromycin selection marker (puro), posttranscriptional regulatory element (WPRE), blasticidin selection marker (blast), and elongation factor-1α promoter (EF1a). (b) Relative functional viral titer of viruses made from lentiCRISPR v1, lentiCRISPR v2, and lentiGuide-Puro vector with a EGFP-targeting sgRNA (n = 3 independently-transfected virus batches with 3 replicate transductions per construct). HEK293FT cells were transduced with serial dilutions of virus and, after 24 hours, selected using puromycin (1 ug/ml). Puromycin-resistant cells were measured after 4 days from the start of selection using the CellTiter-Glo (Promega) luciferase assay. Relative titers were calculated using viral volumes that yielded less than 20% puromycin-resistant cells in order to minimize the number of cells with multiple infections. Numbers above each bar indicate the size of the packaged virus for each construct.
Similar articles
- CRISPR Libraries and Screening.
Poirier JT. Poirier JT. Prog Mol Biol Transl Sci. 2017;152:69-82. doi: 10.1016/bs.pmbts.2017.10.002. Epub 2017 Nov 6. Prog Mol Biol Transl Sci. 2017. PMID: 29150005 - Advances in CRISPR-Cas9 genome engineering: lessons learned from RNA interference.
Barrangou R, Birmingham A, Wiemann S, Beijersbergen RL, Hornung V, Smith Av. Barrangou R, et al. Nucleic Acids Res. 2015 Apr 20;43(7):3407-19. doi: 10.1093/nar/gkv226. Epub 2015 Mar 23. Nucleic Acids Res. 2015. PMID: 25800748 Free PMC article. Review. - CRISPR-Based Lentiviral Knockout Libraries for Functional Genomic Screening and Identification of Phenotype-Related Genes.
Thomsen EA, Mikkelsen JG. Thomsen EA, et al. Methods Mol Biol. 2019;1961:343-357. doi: 10.1007/978-1-4939-9170-9_21. Methods Mol Biol. 2019. PMID: 30912056 - [Advances in application of clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated 9 system in stem cells research].
Sun SJ, Huo JH, Geng ZJ, Sun XY, Fu XB. Sun SJ, et al. Zhonghua Shao Shang Za Zhi. 2018 Apr 20;34(4):253-256. doi: 10.3760/cma.j.issn.1009-2587.2018.04.013. Zhonghua Shao Shang Za Zhi. 2018. PMID: 29690746 Review. Chinese. - Construction of a Set of Novel Transposon Vectors for Efficient Silencing of Protein and lncRNA Genes via CRISPR Interference.
Czarnek M, Kochan J, Wawro M, Myrczek R, Bereta J. Czarnek M, et al. Mol Biotechnol. 2023 Oct;65(10):1598-1607. doi: 10.1007/s12033-023-00675-5. Epub 2023 Jan 28. Mol Biotechnol. 2023. PMID: 36707469 Free PMC article.
Cited by
- Saturation profiling of drug-resistant genetic variants using prime editing.
Kim Y, Oh HC, Lee S, Kim HH. Kim Y, et al. Nat Biotechnol. 2024 Nov 12. doi: 10.1038/s41587-024-02465-z. Online ahead of print. Nat Biotechnol. 2024. PMID: 39533107 - Hepatic SerpinA1 improves energy and glucose metabolism through regulation of preadipocyte proliferation and UCP1 expression.
Okagawa S, Sakaguchi M, Okubo Y, Takekuma Y, Igata M, Kondo T, Takeda N, Araki K, Brandao BB, Qian WJ, Tseng YH, Kulkarni RN, Kubota N, Kahn CR, Araki E. Okagawa S, et al. Nat Commun. 2024 Nov 12;15(1):9585. doi: 10.1038/s41467-024-53835-9. Nat Commun. 2024. PMID: 39532838 - The USP12/46 deubiquitinases protect integrins from ESCRT-mediated lysosomal degradation.
Yu K, Wang GM, Guo SS, Bassermann F, Fässler R. Yu K, et al. EMBO Rep. 2024 Nov 6. doi: 10.1038/s44319-024-00300-9. Online ahead of print. EMBO Rep. 2024. PMID: 39506038 - Design, performance, processing, and validation of a pooled CRISPR perturbation screen for bacterial toxins.
Tian S, Qin Y, Wu Y, Dong M. Tian S, et al. Nat Protoc. 2024 Nov 1. doi: 10.1038/s41596-024-01075-y. Online ahead of print. Nat Protoc. 2024. PMID: 39487259 Review. - A framework for target discovery in rare cancers.
Li B, Sadagopan A, Li J, Wu Y, Cui Y, Konda P, Weiss CN, Choueiri TK, Doench JG, Viswanathan SR. Li B, et al. bioRxiv [Preprint]. 2024 Oct 25:2024.10.24.620074. doi: 10.1101/2024.10.24.620074. bioRxiv. 2024. PMID: 39484513 Free PMC article. Preprint.
References
- Koike-Yusa H, Li Y, Tan E-P, Velasco-Herrera MDC, Yusa K. Nat Biotechnol. 2013;32:267–273. - PubMed
- Zhou Y, et al. Nature. 2014;509:487–491. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials