Oxidative protein folding: from thiol-disulfide exchange reactions to the redox poise of the endoplasmic reticulum - PubMed (original) (raw)
Review
Oxidative protein folding: from thiol-disulfide exchange reactions to the redox poise of the endoplasmic reticulum
Devin A Hudson et al. Free Radic Biol Med. 2015 Mar.
Abstract
This review examines oxidative protein folding within the mammalian endoplasmic reticulum (ER) from an enzymological perspective. In protein disulfide isomerase-first (PDI-first) pathways of oxidative protein folding, PDI is the immediate oxidant of reduced client proteins and then addresses disulfide mispairings in a second isomerization phase. In PDI-second pathways the initial oxidation is PDI-independent. Evidence for the rapid reduction of PDI by reduced glutathione is presented in the context of PDI-first pathways. Strategies and challenges are discussed for determination of the concentrations of reduced and oxidized glutathione and of the ratios of PDI(red):PDI(ox). The preponderance of evidence suggests that the mammalian ER is more reducing than first envisaged. The average redox state of major PDI-family members is largely to almost totally reduced. These observations are consistent with model studies showing that oxidative protein folding proceeds most efficiently at a reducing redox poise consistent with a stoichiometric insertion of disulfides into client proteins. After a discussion of the use of natively encoded fluorescent probes to report the glutathione redox poise of the ER, this review concludes with an elaboration of a complementary strategy to discontinuously survey the redox state of as many redox-active disulfides as can be identified by ratiometric LC-MS-MS methods. Consortia of oxidoreductases that are in redox equilibrium can then be identified and compared to the glutathione redox poise of the ER to gain a more detailed understanding of the factors that influence oxidative protein folding within the secretory compartment.
Keywords: Disulfide exchange; Endoplasmic reticulum; Ero1; Glutathione; Oxidative protein folding; Peroxiredoxin; Protein disulfide isomerase; Quiescin sulfhydryl oxidase; Ratiometric mass spectrometry; Redox potential.
Copyright © 2014 Elsevier Inc. All rights reserved.
Figures
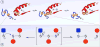
Figure 1. Mechanistic aspects of thiol-disulfide exchange reactions
Panel I illustrates reduction of a CxxC disulfide motif in Aox by an unstructured reduced peptide dithiol (Bred; blue). Generation of the mixed-disulfide intermediate A-B involves an in-line transition state as depicted in the detail shown in Panel II.
Figure 2
Structure of human PDI. The coordinates for the oxidized protein are from Wang et al. [28]. Redox-active CxxC motifs are found in both a and**a′** domains. The N-terminal cysteine sulfur atom of each motif (orange) is solvent accessible and engages in mixed disulfides with redox partners and proteins undergoing disulfide editing. The C-terminal cysteine by contrast is largely buried from solvent (yellow). The b and**b′** domains are redox-inactive. Protein clients of PDI can occupy the central cavity with significant hydrophobic interactions with the**b′** domain.
Figure 3
An example of a PDI-first pathway in oxidative protein folding. In the first phase the unfolded reduced protein is oxidized by PDIox. Here, reduced PDI is regenerated by the FAD-linked sulfhydryl oxidase, Ero1, with the formation of hydrogen peroxide. PDIred is then involved in correction of mispaired disulfides prior to the iterative emergence of the native protein fold.
Figure 4
Some enzymatic and non-enzymatic oxidants for reduced PDI.
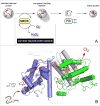
Figure 5
PDI-second pathways of oxidative folding. The initial oxidation of reduced proteins is PDI-independent so that PDI is only engaged in the second phase of oxidative protein folding (Panel A). One facile direct oxidant of reduced conformationally-mobile proteins is Quiescin-sulfhydryl oxidase. Panel B shows the structure of the open form of QSOX from_Trypanosoma brucei_ (3QCP; [52]).
Figure 6
Reduction of PDI by GSH. The model and rate constants of Darby and Creighton [57] were used for the**a** domain of human PDI reduced with glutathione (Panel A). Panel B shows the time course for oxidized, reduced and mixed disulfide forms of the single CxxC motif using 100 μM PDI and 5 mM GSH [59]. Under these conditions the half-time for equilibration is 0.52 sec; t1/2 values for 2.5 mM and 10 mM GSH are 1.12 and 0.25 sec respectively.
Figure 7
A potential pathway for redox cycling in the ER.
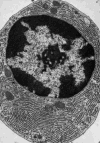
Figure 8
Electron micrograph of a plasma cell from guinea pig bone marrow showing an extensive reticular network of ER. Reproduced from [103].
Figure 9
The use of NEM to trap intraluminal redox state within the ER. The highlighted equilibrium depicts the reduction of one PDI CxxC motif by GSH. Approximately one proton is released at neutral pH values (see the text).
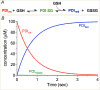
Figure 10
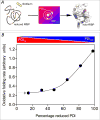
The rate of oxidative refolding of riboflavin binding protein in the presence of PDI redox buffers of defined concentration. Refolding was followed continuously by the quenching of riboflavin binding on association with folded oxidized RfBP in the absence of small molecule redox species and without other enzymatic catalysts of disulfide bond generation (panel A). Rates of refolding are plotted as a function of the percentage of PDIred in the PDI redox buffer (panel B). The concentration of reduced RfBP was 1 μM and the aggregate concentration of reduced and oxidized forms comprising the PDI redox buffer was 30-fold higher (see the Text). Data redrawn from Rancy and Thorpe [49].
Figure 11
Schematic diagram of equilibrating and non-equilibrating redox species in the ER. Redoxactive proteins are distinguished by their shapes; the state of their redox active disulfides is shown by the blue and red shading. Some species are envisaged as directly in equilibrium with the glutathione redox pool and would have a range of redox states depending on their standard redox potentials. Other proteins are represented as interacting indirectly with the glutathione pool or with consortia of other proteins insulated from small molecular weight redox buffers.
Figure 12
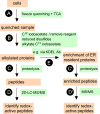
A suggested protocol for a more global analysis of the redox state of thiol-disulfide oxidoreductases within the Er (see the Text).
Similar articles
- Oxidative protein folding in vitro: a study of the cooperation between quiescin-sulfhydryl oxidase and protein disulfide isomerase.
Rancy PC, Thorpe C. Rancy PC, et al. Biochemistry. 2008 Nov 18;47(46):12047-56. doi: 10.1021/bi801604x. Epub 2008 Oct 21. Biochemistry. 2008. PMID: 18937500 Free PMC article. - Novel Roles of the Non-catalytic Elements of Yeast Protein-disulfide Isomerase in Its Interplay with Endoplasmic Reticulum Oxidoreductin 1.
Niu Y, Zhang L, Yu J, Wang CC, Wang L. Niu Y, et al. J Biol Chem. 2016 Apr 8;291(15):8283-94. doi: 10.1074/jbc.M115.694257. Epub 2016 Feb 4. J Biol Chem. 2016. PMID: 26846856 Free PMC article. - Regulation of plant ER oxidoreductin 1 (ERO1) activity for efficient oxidative protein folding.
Matsusaki M, Okuda A, Matsuo K, Gekko K, Masuda T, Naruo Y, Hirose A, Kono K, Tsuchi Y, Urade R. Matsusaki M, et al. J Biol Chem. 2019 Dec 6;294(49):18820-18835. doi: 10.1074/jbc.RA119.010917. Epub 2019 Nov 4. J Biol Chem. 2019. PMID: 31685660 Free PMC article. - Multiple ways to make disulfides.
Bulleid NJ, Ellgaard L. Bulleid NJ, et al. Trends Biochem Sci. 2011 Sep;36(9):485-92. doi: 10.1016/j.tibs.2011.05.004. Epub 2011 Jul 19. Trends Biochem Sci. 2011. PMID: 21778060 Review. - Oxidative protein folding fidelity and redoxtasis in the endoplasmic reticulum.
Wang L, Wang CC. Wang L, et al. Trends Biochem Sci. 2023 Jan;48(1):40-52. doi: 10.1016/j.tibs.2022.06.011. Epub 2022 Jul 20. Trends Biochem Sci. 2023. PMID: 35871147 Review.
Cited by
- Redox Signaling by Reactive Electrophiles and Oxidants.
Parvez S, Long MJC, Poganik JR, Aye Y. Parvez S, et al. Chem Rev. 2018 Sep 26;118(18):8798-8888. doi: 10.1021/acs.chemrev.7b00698. Epub 2018 Aug 27. Chem Rev. 2018. PMID: 30148624 Free PMC article. Review. - Protein Disulfide Isomerases Function as the Missing Link Between Diabetes and Cancer.
Jiang H, Thapa P, Hao Y, Ding N, Alshahrani A, Wei Q. Jiang H, et al. Antioxid Redox Signal. 2022 Dec;37(16-18):1191-1205. doi: 10.1089/ars.2022.0098. Epub 2022 Nov 21. Antioxid Redox Signal. 2022. PMID: 36000195 Free PMC article. Review. - Functions and mechanisms of protein disulfide isomerase family in cancer emergence.
Rahman NSA, Zahari S, Syafruddin SE, Firdaus-Raih M, Low TY, Mohtar MA. Rahman NSA, et al. Cell Biosci. 2022 Aug 14;12(1):129. doi: 10.1186/s13578-022-00868-6. Cell Biosci. 2022. PMID: 35965326 Free PMC article. Review. - Protein disulphide isomerase can predict the clinical prognostic value and contribute to malignant progression in gliomas.
Hu Q, Huang K, Tao C, Zhu X. Hu Q, et al. J Cell Mol Med. 2020 May;24(10):5888-5900. doi: 10.1111/jcmm.15264. Epub 2020 Apr 17. J Cell Mol Med. 2020. PMID: 32301283 Free PMC article. - Machine Learning and Network Analysis of Molecular Dynamics Trajectories Reveal Two Chains of Red/Ox-specific Residue Interactions in Human Protein Disulfide Isomerase.
Karamzadeh R, Karimi-Jafari MH, Sharifi-Zarchi A, Chitsaz H, Salekdeh GH, Moosavi-Movahedi AA. Karamzadeh R, et al. Sci Rep. 2017 Jun 16;7(1):3666. doi: 10.1038/s41598-017-03966-5. Sci Rep. 2017. PMID: 28623339 Free PMC article.
References
- Anfinsen CB. Principles that Govern the Folding of Protein Chains. Science. 1973;181:223–230. - PubMed
- Riemer J, Bulleid N, Herrmann JM. Disulfide formation in the ER and mitochondria: two solutions to a common process. Science. 2009;324:1284–1287. - PubMed
- Hatahet F, Ruddock LW. Protein Disulfide Isomerase: A Critical Evaluation of Its Function in Disulfide Bond Formation. Antioxid Redox Signalling. 2009;11:2807–2850. - PubMed
- Braakman I, Bulleid NJ. Protein folding and modification in the mammalian endoplasmic reticulum. Ann Rev Biochem. 2011;80:71–99. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 GM026643/GM/NIGMS NIH HHS/United States
- T32 GM008550/GM/NIGMS NIH HHS/United States
- 1-T32-GM008550/GM/NIGMS NIH HHS/United States
- GM26643/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources