The clinical significance of small copy number variants in neurodevelopmental disorders - PubMed (original) (raw)
. 2014 Oct;51(10):677-88.
doi: 10.1136/jmedgenet-2014-102588. Epub 2014 Aug 8.
Beatrice Oneda 1, Pascal Joset 1, Silvia Azzarello-Burri 1, Deborah Bartholdi 1, Katharina Steindl 1, Marie Vincent 1, Joana Cobilanschi 1, Heinrich Sticht 2, Rosa Baldinger 1, Regina Reissmann 1, Irene Sudholt 1, Christian T Thiel 3, Arif B Ekici 3, André Reis 3, Emilia K Bijlsma 4, Joris Andrieux 5, Anne Dieux 6, David FitzPatrick 7, Susanne Ritter 8, Alessandra Baumer 1, Beatrice Latal 8, Barbara Plecko 9, Oskar G Jenni 8, Anita Rauch 1
Affiliations
- PMID: 25106414
- PMCID: PMC4173859
- DOI: 10.1136/jmedgenet-2014-102588
Free PMC article
The clinical significance of small copy number variants in neurodevelopmental disorders
Reza Asadollahi et al. J Med Genet. 2014 Oct.
Free PMC article
Abstract
Background: Despite abundant evidence for pathogenicity of large copy number variants (CNVs) in neurodevelopmental disorders (NDDs), the individual significance of genome-wide rare CNVs <500 kb has not been well elucidated in a clinical context.
Methods: By high-resolution chromosomal microarray analysis, we investigated the clinical significance of all rare non-polymorphic exonic CNVs sizing 1-500 kb in a cohort of 714 patients with undiagnosed NDDs.
Results: We detected 96 rare CNVs <500 kb affecting coding regions, of which 58 (60.4%) were confirmed. 6 of 14 confirmed de novo, one of two homozygous and four heterozygous inherited CNVs affected the known microdeletion regions 17q21.31, 16p11.2 and 2p21 or OMIM morbid genes (CASK, CREBBP, PAFAH1B1, SATB2; AUTS2, NRXN3, GRM8). Two further de novo CNVs affecting single genes (MED13L, CTNND2) were instrumental in delineating novel recurrent conditions. For the first time, we here report exonic deletions of CTNND2 causing low normal IQ with learning difficulties with or without autism spectrum disorder. Additionally, we discovered a homozygous out-of-frame deletion of ACOT7 associated with features comparable to the published mouse model. In total, 24.1% of the confirmed small CNVs were categorised as pathogenic or likely pathogenic (median size 130 kb), 17.2% as likely benign, 3.4% represented incidental findings and 55.2% remained unclear.
Conclusions: These results verify the diagnostic relevance of genome-wide rare CNVs <500 kb, which were found pathogenic in ∼2% (14/714) of cases (1.1% de novo, 0.3% homozygous, 0.6% inherited) and highlight their inherent potential for discovery of new conditions.
Keywords: Clinical Genetics; Copy-Number; Developmental; Diagnostics; Genome-Wide.
Published by the BMJ Publishing Group Limited. For permission to use (where not already granted under a licence) please go to http://group.bmj.com/group/rights-licensing/permissions.
Figures
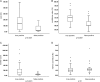
Figure 1
Comparison between true versus false positive status of small copy number variants (CNVs) detected by chromosomal microarray analysis (CMA) and their size, confidence value, marker count and marker count per kb. (A) False positive CNVs were significantly smaller in size (3–181 kb, median 19 kb, mean 45.3 kb) than true CNVs (2–492 kb, median 131 kb, mean 164.7 kb) (p<0.0001). (B) There was also a significant difference between the two groups regarding their confidence values (mean of 88.9% vs 91.9%, p<0.0001) and (C) marker count (median of 20 vs 96, mean of 39.9 vs 163.8, p<0.0001). (D) No significant difference was observed for the marker count per kb within the CNV (1.5±1.14 vs 1.1±0.67, p=0.1).
Figure 2
Schematic representation of CTNND2 deletions detected in patients. Patient 1 (DECIPHER #284528) with a de novo deletion of exons 4–7, patient 2 (DECIPHER #248402) with a paternally inherited deletion encompassing exons 2–8, patient 3 (DECIPHER #269928) with a maternally inherited deletion of exons 1–3 and 5'UTR and patient 4 (DECIPHER #271234) with a maternally inherited deletion of exon 3.
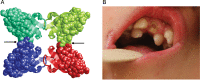
Figure 3
Structure of the Satb2 tetramerisation domain and upper incisors in the patient with intragenic SATB2 duplication. (A) The four subunits of the tetramer are shown in different colours and those parts, which are duplicated in the mutant, are shown in space-filled presentation. This duplication will affect the interfaces between the dimers that form the tetramer (black arrows). Thus, the intragenic duplication is expected to hamper formation of the tetramer, which was suggested to play an important role for long-range chromatin organisation and coordination in gene regulation. (B) Double row of upper incisors in the patient with 32 kb pathogenic duplication within SATB2 at age 3 3/12 years.
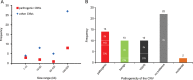
Figure 4
Distribution of copy number variants (CNVs) <500 kb in different size ranges and categories. (A) Frequency of pathogenic or likely pathogenic CNVs (pathogenic) versus other CNVs in four size ranges is shown. (B) Frequency of CNV inheritance pattern in five categories: pathogenic or likely pathogenic (pathogenic), likely benign (benign), variants of uncertain significance (VOUS), CNVs with no evidence in favour or against their pathogenicity (no evidence), and incidental findings related to NDDs (incidental). De novo or likely de novo CNVs are indicated as DN, inherited or likely inherited as IN, and homozygous as HO.
Similar articles
- Burden of Rare Copy Number Variants in Microcephaly: A Brazilian Cohort of 185 Microcephalic Patients and Review of the Literature.
Tolezano GC, Bastos GC, da Costa SS, Freire BL, Homma TK, Honjo RS, Yamamoto GL, Passos-Bueno MR, Koiffmann CP, Kim CA, Vianna-Morgante AM, de Lima Jorge AA, Bertola DR, Rosenberg C, Krepischi ACV. Tolezano GC, et al. J Autism Dev Disord. 2024 Mar;54(3):1181-1212. doi: 10.1007/s10803-022-05853-z. Epub 2022 Dec 11. J Autism Dev Disord. 2024. PMID: 36502452 Review. - The clinical benefit of array-based comparative genomic hybridization for detection of copy number variants in Czech children with intellectual disability and developmental delay.
Wayhelova M, Smetana J, Vallova V, Hladilkova E, Filkova H, Hanakova M, Vilemova M, Nikolova P, Gromesova B, Gaillyova R, Kuglik P. Wayhelova M, et al. BMC Med Genomics. 2019 Jul 23;12(1):111. doi: 10.1186/s12920-019-0559-7. BMC Med Genomics. 2019. PMID: 31337399 Free PMC article. - Identification of novel candidate disease genes from de novo exonic copy number variants.
Gambin T, Yuan B, Bi W, Liu P, Rosenfeld JA, Coban-Akdemir Z, Pursley AN, Nagamani SCS, Marom R, Golla S, Dengle L, Petrie HG, Matalon R, Emrick L, Proud MB, Treadwell-Deering D, Chao HT, Koillinen H, Brown C, Urraca N, Mostafavi R, Bernes S, Roeder ER, Nugent KM, Bader PI, Bellus G, Cummings M, Northrup H, Ashfaq M, Westman R, Wildin R, Beck AE, Immken L, Elton L, Varghese S, Buchanan E, Faivre L, Lefebvre M, Schaaf CP, Walkiewicz M, Yang Y, Kang SL, Lalani SR, Bacino CA, Beaudet AL, Breman AM, Smith JL, Cheung SW, Lupski JR, Patel A, Shaw CA, Stankiewicz P. Gambin T, et al. Genome Med. 2017 Sep 21;9(1):83. doi: 10.1186/s13073-017-0472-7. Genome Med. 2017. PMID: 28934986 Free PMC article. - Both rare and de novo copy number variants are prevalent in agenesis of the corpus callosum but not in cerebellar hypoplasia or polymicrogyria.
Sajan SA, Fernandez L, Nieh SE, Rider E, Bukshpun P, Wakahiro M, Christian SL, Rivière JB, Sullivan CT, Sudi J, Herriges MJ, Paciorkowski AR, Barkovich AJ, Glessner JT, Millen KJ, Hakonarson H, Dobyns WB, Sherr EH. Sajan SA, et al. PLoS Genet. 2013;9(10):e1003823. doi: 10.1371/journal.pgen.1003823. Epub 2013 Oct 3. PLoS Genet. 2013. PMID: 24098143 Free PMC article. - Genomic copy number variation in disorders of cognitive development.
Morrow EM. Morrow EM. J Am Acad Child Adolesc Psychiatry. 2010 Nov;49(11):1091-104. doi: 10.1016/j.jaac.2010.08.009. J Am Acad Child Adolesc Psychiatry. 2010. PMID: 20970697 Free PMC article. Review.
Cited by
- Burden analysis of rare microdeletions suggests a strong impact of neurodevelopmental genes in genetic generalised epilepsies.
Lal D, Ruppert AK, Trucks H, Schulz H, de Kovel CG, Kasteleijn-Nolst Trenité D, Sonsma AC, Koeleman BP, Lindhout D, Weber YG, Lerche H, Kapser C, Schankin CJ, Kunz WS, Surges R, Elger CE, Gaus V, Schmitz B, Helbig I, Muhle H, Stephani U, Klein KM, Rosenow F, Neubauer BA, Reinthaler EM, Zimprich F, Feucht M, Møller RS, Hjalgrim H, De Jonghe P, Suls A, Lieb W, Franke A, Strauch K, Gieger C, Schurmann C, Schminke U, Nürnberg P; EPICURE Consortium; Sander T. Lal D, et al. PLoS Genet. 2015 May 7;11(5):e1005226. doi: 10.1371/journal.pgen.1005226. eCollection 2015 May. PLoS Genet. 2015. PMID: 25950944 Free PMC article. - Burden of Rare Copy Number Variants in Microcephaly: A Brazilian Cohort of 185 Microcephalic Patients and Review of the Literature.
Tolezano GC, Bastos GC, da Costa SS, Freire BL, Homma TK, Honjo RS, Yamamoto GL, Passos-Bueno MR, Koiffmann CP, Kim CA, Vianna-Morgante AM, de Lima Jorge AA, Bertola DR, Rosenberg C, Krepischi ACV. Tolezano GC, et al. J Autism Dev Disord. 2024 Mar;54(3):1181-1212. doi: 10.1007/s10803-022-05853-z. Epub 2022 Dec 11. J Autism Dev Disord. 2024. PMID: 36502452 Review. - A pilot study on glutamate receptor and carrier gene variants and risk of childhood autism spectrum.
Liu J, Yan J, Qu F, Mo W, Yu H, Hu P, Zhang Z. Liu J, et al. Metab Brain Dis. 2023 Oct;38(7):2477-2488. doi: 10.1007/s11011-023-01272-w. Epub 2023 Aug 14. Metab Brain Dis. 2023. PMID: 37578654 - Loss of ACOT7 potentiates seizures and metabolic dysfunction.
Bowman CE, Selen Alpergin ES, Ellis JM, Wolfgang MJ. Bowman CE, et al. Am J Physiol Endocrinol Metab. 2019 Nov 1;317(5):E941-E951. doi: 10.1152/ajpendo.00537.2018. Epub 2019 Apr 30. Am J Physiol Endocrinol Metab. 2019. PMID: 31039008 Free PMC article. - High-resolution chromosomal microarray analysis for copy-number variations in high-functioning autism reveals large aberration typical for intellectual disability.
Werling AM, Grünblatt E, Oneda B, Bobrowski E, Gundelfinger R, Taurines R, Romanos M, Rauch A, Walitza S. Werling AM, et al. J Neural Transm (Vienna). 2020 Jan;127(1):81-94. doi: 10.1007/s00702-019-02114-9. Epub 2019 Dec 14. J Neural Transm (Vienna). 2020. PMID: 31838600
References
- Lee JA, Lupski JR. Genomic rearrangements and gene copy-number alterations as a cause of nervous system disorders. Neuron 2006;52:103–21 - PubMed
- Ropers HH. Genetics of early onset cognitive impairment. Annu Rev Genomics Hum Genet 2010;11:161–87 - PubMed
- Miller DT, Adam MP, Aradhya S, Biesecker LG, Brothman AR, Carter NP, Church DM, Crolla JA, Eichler EE, Epstein CJ, Faucett WA, Feuk L, Friedman JM, Hamosh A, Jackson L, Kaminsky EB, Kok K, Krantz ID, Kuhn RM, Lee C, Ostell JM, Rosenberg C, Scherer SW, Spinner NB, Stavropoulos DJ, Tepperberg JH, Thorland EC, Vermeesch JR, Waggoner DJ, Watson MS, Martin CL, Ledbetter DH. Consensus statement: chromosomal microarray is a first-tier clinical diagnostic test for individuals with developmental disabilities or congenital anomalies. Am J Hum Genet 2010;86:749–64 - PMC - PubMed
- Conrad DF, Pinto D, Redon R, Feuk L, Gokcumen O, Zhang Y, Aerts J, Andrews TD, Barnes C, Campbell P, Fitzgerald T, Hu M, Ihm CH, Kristiansson K, Macarthur DG, Macdonald JR, Onyiah I, Pang AW, Robson S, Stirrups K, Valsesia A, Walter K, Wei J, Tyler-Smith C, Carter NP, Lee C, Scherer SW, Hurles ME. Origins and functional impact of copy number variation in the human genome. Nature 2010;464:704–12 - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous