Proteasome assembly - PubMed (original) (raw)
Review
Proteasome assembly
Zhu Chao Gu et al. Cell Mol Life Sci. 2014 Dec.
Abstract
In eukaryotic cells, proteasomes are highly conserved protease complexes and eliminate unwanted proteins which are marked by poly-ubiquitin chains for degradation. The 26S proteasome consists of the proteolytic core particle, the 20S proteasome, and the 19S regulatory particle, which are composed of 14 and 19 different subunits, respectively. Proteasomes are the second-most abundant protein complexes and are continuously assembled from inactive precursor complexes in proliferating cells. The modular concept of proteasome assembly was recognized in prokaryotic ancestors and applies to eukaryotic successors. The efficiency and fidelity of eukaryotic proteasome assembly is achieved by several proteasome-dedicated chaperones that initiate subunit incorporation and control the quality of proteasome assemblies by transiently interacting with proteasome precursors. It is important to understand the mechanism of proteasome assembly as the proteasome has key functions in the turnover of short-lived proteins regulating diverse biological processes.
Figures
Fig. 1
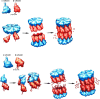
Assembly of archaeal and eubacterial proteasome core particles (CP). The CP consists of α- and β-type subunits. In T. acidophilum, the first step is the assembly of the α ring which serves as template for pro-β subunit incorporation. The dimerization of two half CP yields the pre-holo-CP which is converted into the mature CP by autocatalytic cleavage of the β-propeptides (upper panel). In R. erythropolis, the first step is the formation of an α/pro-β heterodimer. The oligomerization of seven α/pro-β dimers results in the formation of the half-CP. Two half-CP dimerize into the pre-holo-CP in which β-propeptide processing yields CP maturation (lower panel)
Fig. 2
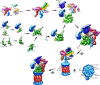
The assembly of eukaryotic CP involves CP-dedicated chaperones. The chaperones PAC1–PAC2 assist the assembly of the α ring from seven different α subunits and prevent faulty α–α ring dimerization. The chaperones PAC3–PAC4 support the incorporation of pro-β2. PAC3–PAC4 is released, when β3, β4 and Ump1 join the α ring. With the incorporation of pro-β1, pro-β5 and pro-β6 the half-CP is almost completed. The dimerization of two half-CP into the pre-holo-CP is stabilized by the incorporation of pro-β7, the last β subunit. The half-CP is also found to be associated with Blm10, especially, if the ability to close the α ring gates with CP maturation is impaired. Within the pre-holo-CP, the β-propeptides are removed by autocatalytic cleavage and Ump is degraded by the nascent CP. PAC1–PAC2, Blm10 and the RP are bound to the nascent and mature CP, if the α ring gates are disordered or opened. Usually, the α ring gates are closed within free mature CP. See main text for details
Fig. 3
The model of CP-independent RP assembly is depicted. In the CP-independent assembly pathway, four RP-dedicated chaperones join RP base modules: Hsm3/S5b binds to the ATPase subunits Rpt1 and Rpt2, Nas2/p27 to Rpt5, Nas6/p28 to Rpt3 and Rpn14/PAAF1 to Rpt6, respectively, by forming three modules or precursors, namely, the Nas6/p28-Rpt3–Rpt6-Rpn14/PPAF1 module, the Nas2/p27-Rpt5–Rpt4 module and the Hsm3/S5b-Rpt1–Rpt2-Rpn2 module. The chaperones are dislocated with the attachment to the CP. The RP lid is assembled from two modules, the Rpn5, 6, 8, 9 and 11 module and the Rpn3, 7 and Sem1 module. The incorporation of Rpn12 completes RP lid assembly. RP base and lid associate with Rpn10 and bind to the CP. Rpn6 also stabilizes the RP–CP interaction. In the CP-dependent assembly pathway, the RP base modules successively dock onto the CP template. The lid modules are assembled on top of the CP–RP base. In the CP-dependent model, the chaperones are most likely released, when the RP base is docking on the CP template to avoid chaperone collisions with the CP–RP base interface. Aberrant RP–CP assemblies are preferentially recognized by Ecm29, which may contact the RP and CP. The structure of Ecm29-bound RP–CP assemblies is unknown. Structures of the proteasomal subunits and proteasomal chaperones were deduced from single particle electron cryo-microscopy and crystal structures deposited in the PDB database. See main text for details
Similar articles
- Chaperone-assisted assembly of the proteasome core particle.
Matias AC, Ramos PC, Dohmen RJ. Matias AC, et al. Biochem Soc Trans. 2010 Feb;38(Pt 1):29-33. doi: 10.1042/BST0380029. Biochem Soc Trans. 2010. PMID: 20074030 - Molecular mechanisms of proteasome assembly.
Murata S, Yashiroda H, Tanaka K. Murata S, et al. Nat Rev Mol Cell Biol. 2009 Feb;10(2):104-15. doi: 10.1038/nrm2630. Nat Rev Mol Cell Biol. 2009. PMID: 19165213 Review. - A heterodimeric complex that promotes the assembly of mammalian 20S proteasomes.
Hirano Y, Hendil KB, Yashiroda H, Iemura S, Nagane R, Hioki Y, Natsume T, Tanaka K, Murata S. Hirano Y, et al. Nature. 2005 Oct 27;437(7063):1381-5. doi: 10.1038/nature04106. Nature. 2005. PMID: 16251969 - Chaperone-driven proteasome assembly.
Rosenzweig R, Glickman MH. Rosenzweig R, et al. Biochem Soc Trans. 2008 Oct;36(Pt 5):807-12. doi: 10.1042/BST0360807. Biochem Soc Trans. 2008. PMID: 18793141 Review. - In-depth Analysis of the Lid Subunits Assembly Mechanism in Mammals.
Bai M, Zhao X, Sahara K, Ohte Y, Hirano Y, Kaneko T, Yashiroda H, Murata S. Bai M, et al. Biomolecules. 2019 May 31;9(6):213. doi: 10.3390/biom9060213. Biomolecules. 2019. PMID: 31159305 Free PMC article.
Cited by
- Proteomic Assessment of Extracellular Vesicles from Canine Tissue Explants as a Pipeline to Identify Molecular Targets in Osteosarcoma: PSMD14/Rpn11 as a Proof of Principle.
Luu AK, Cadieux M, Wong M, Macdonald R, Jones R, Choi D, Oblak M, Brisson B, Sauer S, Chafitz J, Warshawsky D, Wood GA, Viloria-Petit AM. Luu AK, et al. Int J Mol Sci. 2022 Mar 17;23(6):3256. doi: 10.3390/ijms23063256. Int J Mol Sci. 2022. PMID: 35328679 Free PMC article. - Proteomic mapping of Drosophila transgenic elav.L-GAL4/+ brain as a tool to illuminate neuropathology mechanisms.
Velentzas AD, Katarachia SA, Sagioglou NE, Tsioka MM, Anagnostopoulos AK, Mpakou VE, Theotoki EI, Giannopoulou AF, Keramaris KE, Papassideri IS, Tsangaris GT, Stravopodis DJ. Velentzas AD, et al. Sci Rep. 2020 Mar 25;10(1):5430. doi: 10.1038/s41598-020-62510-0. Sci Rep. 2020. PMID: 32214222 Free PMC article. - Bioinformatic and clinical experimental assay uncovers resistance and susceptibility mechanisms of human glioblastomas to temozolomide and identifies new combined and individual survival biomarkers outperforming MGMT promoter methylation.
Modestov A, Zolotovskaia M, Suntsova M, Zakharova G, Seryakov A, Jovcevska I, Mlakar J, Poddubskaya E, Moisseev A, Vykhodtsev G, Roumiantsev S, Sorokin M, Tkachev V, Simonov A, Buzdin A. Modestov A, et al. Ther Adv Med Oncol. 2024 Nov 8;16:17588359241292269. doi: 10.1177/17588359241292269. eCollection 2024. Ther Adv Med Oncol. 2024. PMID: 39525666 Free PMC article. - Dysfunction in protein clearance by the proteasome: impact on autoinflammatory diseases.
Brehm A, Krüger E. Brehm A, et al. Semin Immunopathol. 2015 Jul;37(4):323-33. doi: 10.1007/s00281-015-0486-4. Epub 2015 May 12. Semin Immunopathol. 2015. PMID: 25963519 Review. - Characterization of polyamine metabolism predicts prognosis, immune profile, and therapeutic efficacy in lung adenocarcinoma patients.
Li Z, Wu Y, Yang W, Wang W, Li J, Huang X, Yang Y, Zhang X, Ye X. Li Z, et al. Front Cell Dev Biol. 2024 Apr 8;12:1331759. doi: 10.3389/fcell.2024.1331759. eCollection 2024. Front Cell Dev Biol. 2024. PMID: 38650895 Free PMC article.
References
- Hershko A, Ciechanover A. The ubiquitin system. Annu Rev Biochem. 1998;67:425–479. - PubMed
- Tanaka K. The proteasome: from basic mechanisms to emerging roles. Keio J Med. 2013;62(1):1–12. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources