Patterned progression of bacterial populations in the premature infant gut - PubMed (original) (raw)
Observational Study
. 2014 Aug 26;111(34):12522-7.
doi: 10.1073/pnas.1409497111. Epub 2014 Aug 11.
Barbara B Warner 2, Yanjiao Zhou 3, George M Weinstock 3, Erica Sodergren 3, Carla M Hall-Moore 2, Harold J Stevens 2, William E Bennett Jr 2, Nurmohammad Shaikh 2, Laura A Linneman 2, Julie A Hoffmann 2, Aaron Hamvas 2, Elena Deych 1, Berkley A Shands 1, William D Shannon 4, Phillip I Tarr 5
Affiliations
- PMID: 25114261
- PMCID: PMC4151715
- DOI: 10.1073/pnas.1409497111
Observational Study
Patterned progression of bacterial populations in the premature infant gut
Patricio S La Rosa et al. Proc Natl Acad Sci U S A. 2014.
Erratum in
- Proc Natl Acad Sci U S A. 2014 Dec 2;111(48):17336
Abstract
In the weeks after birth, the gut acquires a nascent microbiome, and starts its transition to bacterial population equilibrium. This early-in-life microbial population quite likely influences later-in-life host biology. However, we know little about the governance of community development: does the gut serve as a passive incubator where the first organisms randomly encountered gain entry and predominate, or is there an orderly progression of members joining the community of bacteria? We used fine interval enumeration of microbes in stools from multiple subjects to answer this question. We demonstrate via 16S rRNA gene pyrosequencing of 922 specimens from 58 subjects that the gut microbiota of premature infants residing in a tightly controlled microbial environment progresses through a choreographed succession of bacterial classes from Bacilli to Gammaproteobacteria to Clostridia, interrupted by abrupt population changes. As infants approach 33-36 wk postconceptional age (corresponding to the third to the twelfth weeks of life depending on gestational age at birth), the gut is well colonized by anaerobes. Antibiotics, vaginal vs. Caesarian birth, diet, and age of the infants when sampled influence the pace, but not the sequence, of progression. Our results suggest that in infants in a microbiologically constrained ecosphere of a neonatal intensive care unit, gut bacterial communities have an overall nonrandom assembly that is punctuated by microbial population abruptions. The possibility that the pace of this assembly depends more on host biology (chiefly gestational age at birth) than identifiable exogenous factors warrants further consideration.
Keywords: mixed model regression analysis; necrotizing enterocolitis; nonmetric multidimensional scaling; prematurity.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
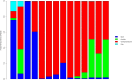
Fig. 1.
Bacterial taxa composition at class level as function of day of life of a single subject. These samples are from subject 32 who was born after 29 wk gestation. Each bar represents the bacterial proportions within single samples. From left to right, bars correspond to stools obtained on days of life 3, 4, 6, 8, 9, 10, 12, 14, 14, 16, 17, 19, 25, and 39, respectively. The predominant classes are Bacilli (dark blue), Gammaproteobacteria (red), and Clostridia (green). Eleven additional classes (light blue) were also present, mostly in stools obtained on days of life 3 and 4.
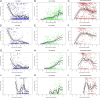
Fig. 2.
Percent bacterial class abundance. Proportion of each bacterial class is represented in y axes in each graph. The x axes represent samples obtained at various postconceptional ages (weeks). The columns represent class proportions, as indicated. (A–C) All infants in cohort (n = 58 subjects). (D–F) Infants in <26 wk gestational age subcohort (_n_ = 15 subjects). (_G–I_) Infants in 26–28 wk gestational age subcohort (_n_ = 20 subjects). (_J–L_) Infants in >28 wk gestational age subcohort (n = 23 subjects). To reduce the effect of random fluctuations in the data (noise), the lines were produced by fitting a cubic smooth spline (thick black line) to the data with 95% confidence bands (thinner lines) generated using a nonparametric bootstrap method.
Comment in
- Gut microbiota: Ordered development of gut microbiota observed in premature infants.
Leake I. Leake I. Nat Rev Gastroenterol Hepatol. 2014 Oct;11(10):579. doi: 10.1038/nrgastro.2014.152. Epub 2014 Aug 26. Nat Rev Gastroenterol Hepatol. 2014. PMID: 25157624 No abstract available.
Similar articles
- The developing premature infant gut microbiome is a major factor shaping the microbiome of neonatal intensive care unit rooms.
Brooks B, Olm MR, Firek BA, Baker R, Geller-McGrath D, Reimer SR, Soenjoyo KR, Yip JS, Dahan D, Thomas BC, Morowitz MJ, Banfield JF. Brooks B, et al. Microbiome. 2018 Jun 20;6(1):112. doi: 10.1186/s40168-018-0493-5. Microbiome. 2018. PMID: 29925423 Free PMC article. - The Microbiome and Metabolome of Preterm Infant Stool Are Personalized and Not Driven by Health Outcomes, Including Necrotizing Enterocolitis and Late-Onset Sepsis.
Wandro S, Osborne S, Enriquez C, Bixby C, Arrieta A, Whiteson K. Wandro S, et al. mSphere. 2018 Jun 6;3(3):e00104-18. doi: 10.1128/mSphere.00104-18. Print 2018 Jun 27. mSphere. 2018. PMID: 29875143 Free PMC article. - Hospitalized Premature Infants Are Colonized by Related Bacterial Strains with Distinct Proteomic Profiles.
Brown CT, Xiong W, Olm MR, Thomas BC, Baker R, Firek B, Morowitz MJ, Hettich RL, Banfield JF. Brown CT, et al. mBio. 2018 Apr 10;9(2):e00441-18. doi: 10.1128/mBio.00441-18. mBio. 2018. PMID: 29636439 Free PMC article. - Bacterial colonization and gut development in preterm neonates.
Cilieborg MS, Boye M, Sangild PT. Cilieborg MS, et al. Early Hum Dev. 2012 Mar;88 Suppl 1:S41-9. doi: 10.1016/j.earlhumdev.2011.12.027. Epub 2012 Jan 28. Early Hum Dev. 2012. PMID: 22284985 Review. - Recent advances in understanding the neonatal microbiome.
Dalby MJ, Hall LJ. Dalby MJ, et al. F1000Res. 2020 May 22;9:F1000 Faculty Rev-422. doi: 10.12688/f1000research.22355.1. eCollection 2020. F1000Res. 2020. PMID: 32518631 Free PMC article. Review.
Cited by
- Necrotizing Enterocolitis and Neurodevelopmental Impairments: Microbiome, Gut, and Brain Entanglements.
Sha C, Jin Z, Ku SY, Kogosov AS, Yu S, Bergese SD, Hsieh H. Sha C, et al. Biomolecules. 2024 Oct 4;14(10):1254. doi: 10.3390/biom14101254. Biomolecules. 2024. PMID: 39456187 Free PMC article. Review. - Longitudinal analysis of the premature infant intestinal microbiome prior to necrotizing enterocolitis: a case-control study.
Zhou Y, Shan G, Sodergren E, Weinstock G, Walker WA, Gregory KE. Zhou Y, et al. PLoS One. 2015 Mar 5;10(3):e0118632. doi: 10.1371/journal.pone.0118632. eCollection 2015. PLoS One. 2015. PMID: 25741698 Free PMC article. - Longitudinal Survey of Microbiota in Hospitalized Preterm Very-Low-Birth-Weight Infants.
Patel AL, Mutlu EA, Sun Y, Koenig L, Green S, Jakubowicz A, Mryan J, Engen P, Fogg L, Chen AL, Pombar X, Meier PP, Keshavarzian A. Patel AL, et al. J Pediatr Gastroenterol Nutr. 2016 Feb;62(2):292-303. doi: 10.1097/MPG.0000000000000913. J Pediatr Gastroenterol Nutr. 2016. PMID: 26230901 Free PMC article. - Antibiotic perturbation of the preterm infant gut microbiome and resistome.
Gasparrini AJ, Crofts TS, Gibson MK, Tarr PI, Warner BB, Dantas G. Gasparrini AJ, et al. Gut Microbes. 2016 Sep 2;7(5):443-9. doi: 10.1080/19490976.2016.1218584. Epub 2016 Jul 29. Gut Microbes. 2016. PMID: 27472377 Free PMC article. Review. - Human milk cream alters intestinal microbiome of preterm infants: a prospective cohort study.
Adeniyi-Ipadeola GO, Hoffman KL, Yang H, Javornik Cregeen SJ, Preidis GA, Ramani S, Hair AB. Adeniyi-Ipadeola GO, et al. Pediatr Res. 2024 May;95(6):1564-1571. doi: 10.1038/s41390-023-02948-w. Epub 2024 Jan 16. Pediatr Res. 2024. PMID: 38228744
References
- Bäckhed F. Host responses to the human microbiome. Nutr Rev. 2012;70(Suppl 1):S14–S17. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- P30DK052574/DK/NIDDK NIH HHS/United States
- U01 HL101465/HL/NHLBI NIH HHS/United States
- UH3AI083265/AI/NIAID NIH HHS/United States
- U54HG004968/HG/NHGRI NIH HHS/United States
- P30 DK052574/DK/NIDDK NIH HHS/United States
- UH3 AI083265/AI/NIAID NIH HHS/United States
- U54 HG004968/HG/NHGRI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials