Local packing density is the main structural determinant of the rate of protein sequence evolution at site level - PubMed (original) (raw)
Local packing density is the main structural determinant of the rate of protein sequence evolution at site level
So-Wei Yeh et al. Biomed Res Int. 2014.
Abstract
Functional and biophysical constraints result in site-dependent patterns of protein sequence variability. It is commonly assumed that the key structural determinant of site-specific rates of evolution is the Relative Solvent Accessibility (RSA). However, a recent study found that amino acid substitution rates correlate better with two Local Packing Density (LPD) measures, the Weighted Contact Number (WCN) and the Contact Number (CN), than with RSA. This work aims at a more thorough assessment. To this end, in addition to substitution rates, we considered four other sequence variability scores, four measures of solvent accessibility (SA), and other CN measures. We compared all properties for each protein of a structurally and functionally diverse representative dataset of monomeric enzymes. We show that the best sequence variability measures take into account phylogenetic tree topology. More importantly, we show that both LPD measures (WCN and CN) correlate better than all of the SA measures, regardless of the sequence variability score used. Moreover, the independent contribution of the best LPD measure is approximately four times larger than that of the best SA measure. This study strongly supports the conclusion that a site's packing density rather than its solvent accessibility is the main structural determinant of its rate of evolution.
Figures
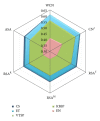
Figure 1
Comparison of sequence variability profiles by their average Pearson's correlation coefficients with different structural profiles. The sequence variability scores (listed in the figure legend) are ConSurf rate of evolution (CS), Evolutionary Trace score (ET), Karlin & Brocchieri Sum-of-Pairs score (KBSP), Valdar & Thornton Sum-of-Pairs score (VTSP), and Entropy (EN). The structural properties (the apices of the hexagon) are Weighted Contact Number (WCN), Contact Number (CN), Relative Solvent Accessibility (RSA), and Absolute Solvent Accessibility (ASA). The asterisk mark on CN means that the cut-off radius was chosen to maximize each CN-sequence average correlation. The cut-off radii for CS, ET, KBSP, VTSP, and EN are 19 Å, 19 Å, 18 Å, 18 Å, and 20 Å, respectively. Superscript letters distinguish RSA profiles obtained using different methods.
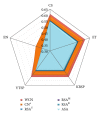
Figure 2
Comparison of structural profiles by their average Pearson's correlation coefficients with different sequence variability profiles. The sequence variability measures (the axes of the pentagon) are ConSurf rate of evolution (CS), Evolutionary Trace score (ET), Karlin & Brocchieri Sum-of-Pairs score (KBSP), Valdar & Thornton Sum-of-Pairs score (VTSP), and Entropy (EN). The structural properties (listed in the figure legend) are Weighted Contact Number (WCN), Contact Number (CN), Relative Solvent Accessibility (RSA), and Absolute Solvent Accessibility (ASA). The asterisk mark on CN means that the cut-off radius was chosen to maximize each CN-sequence average correlation. The cut-off radii for CS, ET, KBSP, VTSP, and EN are 19 Å, 19 Å, 18 Å, 18 Å, and 20 Å, respectively. Superscript letters distinguish RSA profiles obtained using different methods.
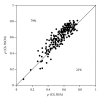
Figure 3
Local packing density versus solvent accessibility as determinants of site-specific evolutionary rates. Points above (below) the diagonal are proteins for which WCN (RSAT) correlates better than RSAT (WCN) with the site-specific rates of amino acid substitution as estimated using the phylogenetic-based approach ConSurf (CS). The percentages of points above and below the diagonals are shown.
Similar articles
- Site-specific structural constraints on protein sequence evolutionary divergence: local packing density versus solvent exposure.
Yeh SW, Liu JW, Yu SH, Shih CH, Hwang JK, Echave J. Yeh SW, et al. Mol Biol Evol. 2014 Jan;31(1):135-9. doi: 10.1093/molbev/mst178. Epub 2013 Oct 8. Mol Biol Evol. 2014. PMID: 24109601 - Too packed to change: side-chain packing and site-specific substitution rates in protein evolution.
Marcos ML, Echave J. Marcos ML, et al. PeerJ. 2015 Apr 23;3:e911. doi: 10.7717/peerj.911. eCollection 2015. PeerJ. 2015. PMID: 25922797 Free PMC article. - A mechanistic stress model of protein evolution accounts for site-specific evolutionary rates and their relationship with packing density and flexibility.
Huang TT, del Valle Marcos ML, Hwang JK, Echave J. Huang TT, et al. BMC Evol Biol. 2014 Apr 9;14:78. doi: 10.1186/1471-2148-14-78. BMC Evol Biol. 2014. PMID: 24716445 Free PMC article. - Structural and functional restraints in the evolution of protein families and superfamilies.
Gong S, Worth CL, Bickerton GR, Lee S, Tanramluk D, Blundell TL. Gong S, et al. Biochem Soc Trans. 2009 Aug;37(Pt 4):727-33. doi: 10.1042/BST0370727. Biochem Soc Trans. 2009. PMID: 19614584 Review. - Structural and functional constraints in the evolution of protein families.
Worth CL, Gong S, Blundell TL. Worth CL, et al. Nat Rev Mol Cell Biol. 2009 Oct;10(10):709-20. doi: 10.1038/nrm2762. Epub 2009 Sep 16. Nat Rev Mol Cell Biol. 2009. PMID: 19756040 Review.
Cited by
- Predicting evolutionary site variability from structure in viral proteins: buriedness, packing, flexibility, and design.
Shahmoradi A, Sydykova DK, Spielman SJ, Jackson EL, Dawson ET, Meyer AG, Wilke CO. Shahmoradi A, et al. J Mol Evol. 2014 Oct;79(3-4):130-42. doi: 10.1007/s00239-014-9644-x. Epub 2014 Sep 13. J Mol Evol. 2014. PMID: 25217382 Free PMC article. - Causes of evolutionary rate variation among protein sites.
Echave J, Spielman SJ, Wilke CO. Echave J, et al. Nat Rev Genet. 2016 Feb;17(2):109-21. doi: 10.1038/nrg.2015.18. Epub 2016 Jan 19. Nat Rev Genet. 2016. PMID: 26781812 Free PMC article. Review. - Dissecting the roles of local packing density and longer-range effects in protein sequence evolution.
Shahmoradi A, Wilke CO. Shahmoradi A, et al. Proteins. 2016 Jun;84(6):841-54. doi: 10.1002/prot.25034. Epub 2016 Apr 9. Proteins. 2016. PMID: 26990194 Free PMC article. - Quantifying evolutionary importance of protein sites: A Tale of two measures.
Sharir-Ivry A, Xia Y. Sharir-Ivry A, et al. PLoS Genet. 2021 Apr 7;17(4):e1009476. doi: 10.1371/journal.pgen.1009476. eCollection 2021 Apr. PLoS Genet. 2021. PMID: 33826605 Free PMC article. - Beyond Thermodynamic Constraints: Evolutionary Sampling Generates Realistic Protein Sequence Variation.
Jiang Q, Teufel AI, Jackson EL, Wilke CO. Jiang Q, et al. Genetics. 2018 Apr;208(4):1387-1395. doi: 10.1534/genetics.118.300699. Epub 2018 Jan 30. Genetics. 2018. PMID: 29382650 Free PMC article.
References
- Pál C, Papp B, Lercher MJ. An integrated view of protein evolution. Nature Reviews Genetics. 2006;7(5):337–348. - PubMed
- Thorne JL. Protein evolution constraints and model-based techniques to study them. Current Opinion in Structural Biology. 2007;17(3):337–341. - PubMed
- Worth CL, Gong S, Blundell TL. Structural and functional constraints in the evolution of protein families. Nature Reviews Molecular Cell Biology. 2009;10(10):709–720. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources