Patterns of admixture and population structure in native populations of Northwest North America - PubMed (original) (raw)
. 2014 Aug 14;10(8):e1004530.
doi: 10.1371/journal.pgen.1004530. eCollection 2014 Aug.
Trevor J Pemberton 2, Romain Laurent 1, Brian M Kemp 3, Angelica Gonzalez-Oliver 4, Clara Gorodezky 5, Cris E Hughes 6, Milena R Shattuck 7, Barbara Petzelt 8, Joycelynn Mitchell 8, Harold Harry 9, Theresa William 10, Rosita Worl 11, Jerome S Cybulski 12, Noah A Rosenberg 13, Ripan S Malhi 14
Affiliations
- PMID: 25122539
- PMCID: PMC4133047
- DOI: 10.1371/journal.pgen.1004530
Patterns of admixture and population structure in native populations of Northwest North America
Paul Verdu et al. PLoS Genet. 2014.
Abstract
The initial contact of European populations with indigenous populations of the Americas produced diverse admixture processes across North, Central, and South America. Recent studies have examined the genetic structure of indigenous populations of Latin America and the Caribbean and their admixed descendants, reporting on the genomic impact of the history of admixture with colonizing populations of European and African ancestry. However, relatively little genomic research has been conducted on admixture in indigenous North American populations. In this study, we analyze genomic data at 475,109 single-nucleotide polymorphisms sampled in indigenous peoples of the Pacific Northwest in British Columbia and Southeast Alaska, populations with a well-documented history of contact with European and Asian traders, fishermen, and contract laborers. We find that the indigenous populations of the Pacific Northwest have higher gene diversity than Latin American indigenous populations. Among the Pacific Northwest populations, interior groups provide more evidence for East Asian admixture, whereas coastal groups have higher levels of European admixture. In contrast with many Latin American indigenous populations, the variance of admixture is high in each of the Pacific Northwest indigenous populations, as expected for recent and ongoing admixture processes. The results reveal some similarities but notable differences between admixture patterns in the Pacific Northwest and those in Latin America, contributing to a more detailed understanding of the genomic consequences of European colonization events throughout the Americas.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
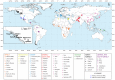
Figure 1. Map of populations included in the combined dataset.
The Tlingit, Tsimshian, Nisga'a, Splatsin, Stswecem'c, and Haida populations, as well as the Northern Mexico Seri population indicated by a black diamond, were newly genotyped for this study. See Tables 1 and S1 for additional population information.
Figure 2. Genome-wide haplotype heterozygosities.
(A) Mean expected haplotype heterozygosity in each population, with standard deviations across the 22 autosomes. (B) The correlation between mean haplotype heterozygosity and geographic distance from Addis Ababa. Population colors and symbols follow Figure 1.
Figure 3. Population-level population structure for 71 populations.
Shown are a (A) multidimensional scaling (MDS) plot (Spearman ρ = 0.965; P<10−15, comparing population-pairwise Euclidean distances on the two-dimensional MDS plot to their population pairwise F ST values), and (B) Neighbor-joining tree based on population pairwise F ST. All the edges of the tree were supported by 100% of the 1,000 bootstrap replicates performed except for four edges corresponding respectively to the Kalash, Caucasian (CEU), Orcadian, and French populations (supported by 76%, 75%, 75%, and 74%, respectively, of the 1,000 bootstrap replicates). Population colors and symbols follow Figure 1.
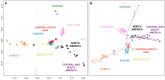
Figure 4. Individual-level population structure.
(A) Multidimensional scaling plot of pairwise allele-sharing distance (ASD) among 2,140 individuals in the combined dataset. (B) Multidimensional scaling plot of pairwise ASD among 528 individuals from 63 worldwide populations, following the resampling of a maximum of 82 individuals each from 11 different population groups. Group choices for resampling were taken from Figure S6. Population colors and symbols follow Figure 1.
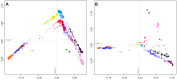
Figure 5. Procrustes-transformed multidimensional scaling plots of Eurasian and American individuals.
(A) 641 individuals from 53 populations after resampling of 82 individuals from each of 14 nonoverlapping groups of European, Central and South Asian, East Asian, and American populations (Figure S7). (B) 641 individuals from 41 populations after resampling of 82 individuals from each of 15 nonoverlapping groups of European, East Asian, and American populations (Figure S8). (C) 393 individuals from 22 populations after resampling of 82 individuals from each of 10 nonoverlapping groups of European and American populations (Figure S9). (D) 450 individuals from 34 populations after resampling of 82 individuals from each of 11 nonoverlapping groups of East Asian and American populations (Figure S10). Procrustes similarity statistics are t 0 = 0.958 between Figures 4B and 5A, t 0 = 0.999 between Figures 5A and 5B, t 0 = 0.956 between Figures 5B and 5C, and t 0 = 0.997 between Figures 5B and 5D. Population colors and symbols follow Figure 1.
Figure 6. Procrustes-transformed multidimensional scaling plot of Pacific Northwest individuals.
The plot is based on pairwise ASD among 82 individuals from six indigenous populations; t 0 = 0.874 between overlapping individuals in Figures 5C and 6. Population colors and symbols follow Figure 1.
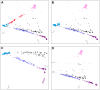
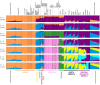
Figure 7. Worldwide Admixture structure.
Plotted are modes with clustering solutions obtained with 30 replicates at each value of K. Values of K and the number of runs in the mode shown appear on the left. In each plot, each cluster is represented by a different color, and each individual is represented by a vertical line divided into K colored segments with heights proportional to genotype memberships in the clusters. Thin black lines separate individuals from different populations. The same 528 individuals included in Figure 4B are considered in the A
dmixture
analyses. Alternate clustering solutions for values of K from 2 to 12 appear in Figure S3.
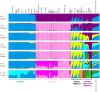
Figure 8. Admixture structure in the Americas.
Plots are as described in Figure 7. The same 641 individuals from 41 European, East Asian, and American populations included in Figure 5B are considered in the A
dmixture
analyses. Alternate clustering solutions for values of K from 2 to 12 appear in Figures S4.
Similar articles
- Genomic Insights into the Ancestry and Demographic History of South America.
Homburger JR, Moreno-Estrada A, Gignoux CR, Nelson D, Sanchez E, Ortiz-Tello P, Pons-Estel BA, Acevedo-Vasquez E, Miranda P, Langefeld CD, Gravel S, Alarcón-Riquelme ME, Bustamante CD. Homburger JR, et al. PLoS Genet. 2015 Dec 4;11(12):e1005602. doi: 10.1371/journal.pgen.1005602. eCollection 2015 Dec. PLoS Genet. 2015. PMID: 26636962 Free PMC article. - Evaluation of group genetic ancestry of populations from Philadelphia and Dakar in the context of sex-biased admixture in the Americas.
Stefflova K, Dulik MC, Pai AA, Walker AH, Zeigler-Johnson CM, Gueye SM, Schurr TG, Rebbeck TR. Stefflova K, et al. PLoS One. 2009 Nov 25;4(11):e7842. doi: 10.1371/journal.pone.0007842. PLoS One. 2009. PMID: 19946364 Free PMC article. - Unravelling the hidden ancestry of American admixed populations.
Montinaro F, Busby GB, Pascali VL, Myers S, Hellenthal G, Capelli C. Montinaro F, et al. Nat Commun. 2015 Mar 24;6:6596. doi: 10.1038/ncomms7596. Nat Commun. 2015. PMID: 25803618 Free PMC article. - A review of ancestrality and admixture in Latin America and the caribbean focusing on native American and African descendant populations.
De Oliveira TC, Secolin R, Lopes-Cendes I. De Oliveira TC, et al. Front Genet. 2023 Jan 19;14:1091269. doi: 10.3389/fgene.2023.1091269. eCollection 2023. Front Genet. 2023. PMID: 36741309 Free PMC article. Review. - Patterns of mtDNA diversity in northwestern North America.
Malhi RS, Breece KE, Shook BA, Kaestle FA, Chatters JC, Hackenberger S, Smith DG. Malhi RS, et al. Hum Biol. 2004 Feb;76(1):33-54. doi: 10.1353/hub.2004.0023. Hum Biol. 2004. PMID: 15222679 Review.
Cited by
- Local Ancestry Inference Based on Population-Specific Single-Nucleotide Polymorphisms-A Study of Admixed Populations in the 1000 Genomes Project.
Fu H, Shi G. Fu H, et al. Genes (Basel). 2024 Aug 21;15(8):1099. doi: 10.3390/genes15081099. Genes (Basel). 2024. PMID: 39202458 Free PMC article. - Ancient DNA uncovers past migrations in California.
Izarraras-Gomez A, Ortega-Del Vecchyo D. Izarraras-Gomez A, et al. Nature. 2023 Dec;624(7990):43-44. doi: 10.1038/d41586-023-03503-7. Nature. 2023. PMID: 37993617 No abstract available. - Modeling the effects of consanguinity on autosomal and X-chromosomal runs of homozygosity and identity-by-descent sharing.
Cotter DJ, Severson AL, Kang JTL, Godrej HN, Carmi S, Rosenberg NA. Cotter DJ, et al. G3 (Bethesda). 2024 Feb 7;14(2):jkad264. doi: 10.1093/g3journal/jkad264. G3 (Bethesda). 2024. PMID: 37972246 Free PMC article. - On the number of genealogical ancestors tracing to the source groups of an admixed population.
Mooney JA, Agranat-Tamir L, Pritchard JK, Rosenberg NA. Mooney JA, et al. Genetics. 2023 Jul 6;224(3):iyad079. doi: 10.1093/genetics/iyad079. Genetics. 2023. PMID: 37410594 Free PMC article. - A paleogenome from a Holocene individual supports genetic continuity in Southeast Alaska.
Aqil A, Gill S, Gokcumen O, Malhi RS, Reese EA, Smith JL, Heaton TT, Lindqvist C. Aqil A, et al. iScience. 2023 Apr 8;26(5):106581. doi: 10.1016/j.isci.2023.106581. eCollection 2023 May 19. iScience. 2023. PMID: 37138779 Free PMC article.
References
- Goebel T, Waters MR, O'Rourke DH (2008) The late Pleistocene dispersal of modern humans in the Americas. Science 319: 1497–1502. - PubMed
- Kemp BM, Schurr TG (2010) Ancient and Modern Genetic Variation in the Americas. In: Auerbach BM, editor. Human variation in the Americas : the integration of archaeology and biological anthropology. Carbondale (IL): Southern Illinois University. pp. 12–50.
- Meltzer DJ (2009) First Peoples in the New World: Colonizing Ice Age America. Berkley: University of California Press. 464 p.
- O'Rourke DH, Raff JA (2010) The human genetic history of the Americas: the final frontier. Current Biology 20: 202–207. - PubMed
- Campbell L (1997) American Indian languages : the historical linguistics of Native America. New York: Oxford University Press. xi, 512 p.
Publication types
MeSH terms
Substances
Grants and funding
This work was supported by US National Science Foundation grants BCS-1025139 and BCS-1147534. AGO was funded in part by the Mexican Consejo Nacional de Ciencia y Tecnología No. 101791. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
LinkOut - more resources
Full Text Sources
Other Literature Sources