Beam-induced motion correction for sub-megadalton cryo-EM particles - PubMed (original) (raw)
Beam-induced motion correction for sub-megadalton cryo-EM particles
Sjors Hw Scheres. Elife. 2014.
Abstract
In electron cryo-microscopy (cryo-EM), the electron beam that is used for imaging also causes the sample to move. This motion blurs the images and limits the resolution attainable by single-particle analysis. In a previous Research article (Bai et al., 2013) we showed that correcting for this motion by processing movies from fast direct-electron detectors allowed structure determination to near-atomic resolution from 35,000 ribosome particles. In this Research advance article, we show that an improved movie processing algorithm is applicable to a much wider range of specimens. The new algorithm estimates straight movement tracks by considering multiple particles that are close to each other in the field of view, and models the fall-off of high-resolution information content by radiation damage in a dose-dependent manner. Application of the new algorithm to four data sets illustrates its potential for significantly improving cryo-EM structures, even for particles that are smaller than 200 kDa.
Keywords: cryo-EM; image analysis; single-particle analysis.
Copyright © 2014, Scheres.
Conflict of interest statement
SHWS: Reviewing editor, eLife.
Figures
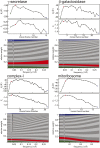
Figure 1.. Beam-induced movement tracks.
A representative micrograph for each of the four test cases is shown, on top of which 50-fold exaggerated beam-induced particle movements are plotted. The original tracks as estimated for running averages of several movie frames for each particle independently are shown in blue; the fitted linear tracks are shown in white. The start and end points of the fitted tracks are indicated with green and red dots, respectively. The orange circles indicate the 2σNB distance for one of the particles on the micrographs. Note that tracks are only shown for those particles that were selected for the final reconstruction after 2D and 3D classification. Also note that the relatively small movement tracks for γ-secretase only represent the beam-induced motion that was not already corrected for in the algorithm by Li et al. (2013). DOI:
http://dx.doi.org/10.7554/eLife.03665.002
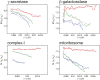
Figure 2.. Radiation-damage weighting.
For each of the four test cases, estimated values for B f and C f (top) and the resulting frequency-dependent relative weights (bottom) are shown for all movie frames. The first, third, and last movie frames of each data set are highlighted in green, red, and blue, respectively. For these movie frames, the relative Guinier plots as described in the main text and the linear fits through them are shown in Figure 2—figure supplement 1. For example, in the γ-secretase case, the third movie frame has the least negative relative B-factor (B f), and therefore this frame contributes the most of all movie frames to the weighted average at the high frequencies (and hence the red band gets broader towards the right-hand side of the relative-weight figure). In contrast, the first and last movie frames have much larger negative B-factors because they suffer from large initial beam-induced motion and radiation damage, respectively. Therefore, these movie frames contribute relatively little to the weighted average at the higher frequencies (and hence the green and blue bands decrease in width towards the right-hand side of the relative-weight figure). Because beam-induced motion and radiation damage affect the low frequencies to a much smaller extent, for the low frequencies all movie frames contribute more or less equally to the weighted average. Therefore, each band is more or less the same width on the left-hand side of the relative-weight figure, although the exact relative weights are dominated by C f on this side of the plot. DOI:
http://dx.doi.org/10.7554/eLife.03665.004
Figure 2—figure supplement 1.. Relative Guinier plots (solid lines) and the linear fits through those (dashed lines) for the first, third, and last movie frames of each data set in green, red, and blue, respectively.
DOI:
http://dx.doi.org/10.7554/eLife.03665.005
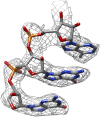
Figure 3.. Map improvement.
Representative parts of the density maps for all four test cases before (left of the arrow) and after the new movie processing approach (right of the arrow). DOI:
http://dx.doi.org/10.7554/eLife.03665.006
Figure 3—figure supplement 1.. The same part of the mitoribosome large sub-unit map as shown in Figure 3, but after application of the original movie processing approach, as described in Bai et al. (2013).
DOI:
http://dx.doi.org/10.7554/eLife.03665.007
Comment in
- Cryo-EM enters a new era.
Kühlbrandt W. Kühlbrandt W. Elife. 2014 Aug 13;3:e03678. doi: 10.7554/eLife.03678. Elife. 2014. PMID: 25122623 Free PMC article.
Similar articles
- Cryo-EM enters a new era.
Kühlbrandt W. Kühlbrandt W. Elife. 2014 Aug 13;3:e03678. doi: 10.7554/eLife.03678. Elife. 2014. PMID: 25122623 Free PMC article. - Ribosome structures to near-atomic resolution from thirty thousand cryo-EM particles.
Bai XC, Fernandez IS, McMullan G, Scheres SH. Bai XC, et al. Elife. 2013 Feb 19;2:e00461. doi: 10.7554/eLife.00461. Elife. 2013. PMID: 23427024 Free PMC article. - A Super-Clustering Approach for Fully Automated Single Particle Picking in Cryo-EM.
Al-Azzawi A, Ouadou A, Tanner JJ, Cheng J. Al-Azzawi A, et al. Genes (Basel). 2019 Aug 30;10(9):666. doi: 10.3390/genes10090666. Genes (Basel). 2019. PMID: 31480377 Free PMC article. - Processing of Structurally Heterogeneous Cryo-EM Data in RELION.
Scheres SH. Scheres SH. Methods Enzymol. 2016;579:125-57. doi: 10.1016/bs.mie.2016.04.012. Epub 2016 May 31. Methods Enzymol. 2016. PMID: 27572726 Review. - Single-particle reconstruction statistics: a diagnostic tool in solving biomolecular structures by cryo-EM.
Heymann JB. Heymann JB. Acta Crystallogr F Struct Biol Commun. 2019 Jan 1;75(Pt 1):33-44. doi: 10.1107/S2053230X18017636. Epub 2019 Jan 1. Acta Crystallogr F Struct Biol Commun. 2019. PMID: 30605123 Free PMC article. Review.
Cited by
- Cryo-EM studies of the structure and dynamics of vacuolar-type ATPases.
Mazhab-Jafari MT, Rubinstein JL. Mazhab-Jafari MT, et al. Sci Adv. 2016 Jul 22;2(7):e1600725. doi: 10.1126/sciadv.1600725. eCollection 2016 Jul. Sci Adv. 2016. PMID: 27532044 Free PMC article. Review. - Analysis and comparison of electron radiation damage assessments in Cryo-EM by single particle analysis and micro-crystal electron diffraction.
Shi D, Huang R. Shi D, et al. Front Mol Biosci. 2022 Oct 5;9:988928. doi: 10.3389/fmolb.2022.988928. eCollection 2022. Front Mol Biosci. 2022. PMID: 36275612 Free PMC article. Review. - 2.8 Å resolution reconstruction of the Thermoplasma acidophilum 20S proteasome using cryo-electron microscopy.
Campbell MG, Veesler D, Cheng A, Potter CS, Carragher B. Campbell MG, et al. Elife. 2015 Mar 11;4:e06380. doi: 10.7554/eLife.06380. Elife. 2015. PMID: 25760083 Free PMC article. - A Bayesian approach to single-particle electron cryo-tomography in RELION-4.0.
Zivanov J, Otón J, Ke Z, von Kügelgen A, Pyle E, Qu K, Morado D, Castaño-Díez D, Zanetti G, Bharat TAM, Briggs JAG, Scheres SHW. Zivanov J, et al. Elife. 2022 Dec 5;11:e83724. doi: 10.7554/eLife.83724. Elife. 2022. PMID: 36468689 Free PMC article. - Glycan shield and epitope masking of a coronavirus spike protein observed by cryo-electron microscopy.
Walls AC, Tortorici MA, Frenz B, Snijder J, Li W, Rey FA, DiMaio F, Bosch BJ, Veesler D. Walls AC, et al. Nat Struct Mol Biol. 2016 Oct;23(10):899-905. doi: 10.1038/nsmb.3293. Epub 2016 Sep 12. Nat Struct Mol Biol. 2016. PMID: 27617430 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources