The CRISPR/Cas9 System Facilitates Clearance of the Intrahepatic HBV Templates In Vivo - PubMed (original) (raw)
doi: 10.1038/mtna.2014.38.
Hung-Chih Yang 2, Yi-Ting Kuo 1, Chun-Jen Liu 3, Ta-Yu Yang 1, Ku-Chun Sung 1, You-Yu Lin 4, Hurng-Yi Wang 5, Chih-Chiang Wang 5, Yueh-Chi Shen 1, Fang-Yi Wu 1, Jia-Horng Kao 6, Ding-Shinn Chen 6, Pei-Jer Chen 6
Affiliations
- PMID: 25137139
- PMCID: PMC4221598
- DOI: 10.1038/mtna.2014.38
The CRISPR/Cas9 System Facilitates Clearance of the Intrahepatic HBV Templates In Vivo
Su-Ru Lin et al. Mol Ther Nucleic Acids. 2014.
Abstract
Persistence of hepatitis B virus (HBV) covalently closed circular DNA (cccDNA) under current antiviral therapy is a major barrier to eradication of chronic hepatitis B (CHB). Curing CHB will require novel strategies for specific disruption of cccDNA. The clustered regularly interspaced short palindromic repeats (CRISPR)/Cas9 system is a newly developed tool for site-specific cleavage of DNA targets directed by a synthetic guide RNA (gRNA) base-paired to the target DNA sequence. To examine whether this system can cleave HBV genomes, we designed eight gRNAs against HBV of genotype A. With the HBV-specific gRNAs, the CRISPR/Cas9 system significantly reduced the production of HBV core and surface proteins in Huh-7 cells transfected with an HBV-expression vector. Among eight screened gRNAs, two effective ones were identified. Interestingly, one gRNA targeting the conserved HBV sequence acted against different genotypes. Using a hydrodynamics-HBV persistence mouse model, we further demonstrated that this system could cleave the intrahepatic HBV genome-containing plasmid and facilitate its clearance in vivo, resulting in reduction of serum surface antigen levels. These data suggest that the CRISPR/Cas9 system could disrupt the HBV-expressing templates both in vitro and in vivo, indicating its potential in eradicating persistent HBV infection.
Figures
Figure 1
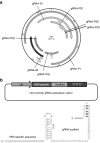
Designs of gRNAs targeting (hepatitis B virus) HBV-specific sequences. (a) Illustration of the gRNA-targeted sequences located in the HBV genome. (b) Designs of the chimeric HBV-specific gRNAs.
Figure 2
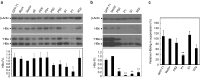
Examination of the efficacy of individual (hepatitis B virus) HBV-specific gRNAs in inhibition of HBV protein expression. The HBV-expression vector was cotransfected to Huh7 cells with (a) each individual gRNA-expression vector and the hCas9 vector or (b) each individual gRNA/Cas9 dual expression vector. The lysate was collected after 48 hours and the levels of intracellular HBcAg and HBsAg were analyzed by using Western blot analysis and quantification of the band intensity. (c) Levels of HBsAg in culture supernatant. The gRNA targeting GFP (GFP-T1), backbone plasmid for the construction of gRNA-expression cassette (blunt), and plasmid containing the U6 promoter/gRNA scaffold (vector) served as negative controls. The bands of *HBs and HBs represent glycosylated and nonglycosylated small surface antigens of HBV, respectively. The relative intensity of HBsAg was shown by quantification of both bands of *HBs and HBs normalized to that of Vector in the bottom panel. RI, relative intensity. The RI and percentages of HBsAg expression were calculated from the results of three independent experiments and presented as mean ± SEM. Statistical significance was computed by Student's _t_-test and indicated as asterisks (*P < 0.05, **P < 0.01).
Figure 3
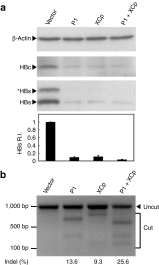
Suppression of the (hepatitis B virus) HBV protein expression via the multiplex HBV-specific gRNA. The HBV-expression vector was cotransfected to Huh7 cells with the gRNA/Cas9 dual expression vectors. The lysate was collected after 48 hours and the levels of intracellular HBsAg were analyzed using Western blotting as shown by this representative figure. The band intensities were quantified by software ImageJ. The relative HBsAg intensity (HBs R.I.) in the bar graph of the bottom panel was the results of four independent experiments and presented as mean ± SEM. (b) The DNA extracted from the transfected Huh7 cells in panel (a) was analyzed by T7E1 assay for determining the percentage of Indel as shown in the bottom.
Figure 4
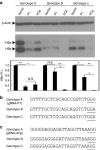
Effects of the gRNAs P1 and XCp on different genotypes of (hepatitis B virus) HBV. (a) HBV-expression vector was cotransfected to Huh7 cells with either the gRNA-P1/Cas9 or gRNA-XCp/Cas9 dual expression vector alone or in combination. The lysate was collected 48 hours post-transfection. The levels of intracellular HBsAg were determined by Western blot analysis. The band intensity was measured by software ImageJ and normalized to the intensity of the mock vector (Vector) for each HBV genotype. The HBs R.I. was calculated from the results of three independent experiments and presented as mean ± SEM. Statistical significance was calculated by the Student's _t-_test and indicated by asterisks (*P < 0.05, **P < 0.01). N.S. means no statistical significance. Sequence alignment of gRNAs P1 (b) and XCp (c) in HBV genotypes A, B, and C. The underlined nucleotides indicated the PAM motif for the recognition of Cas9.
Figure 5
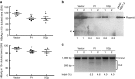
In vivo destruction of intrahepatic (hepatitis B virus) HBV genomes by the HBV-specific CRISPR/Cas9 system. Sera from mice receiving pAAV/HBV 1.2 together with the gRNA/Cas9 dull expression plasmid, vector (n = 4), gRNA P1 (n = 4), or gRNA XCp (n = 4), were collected after hydrodynamic injection. (a) Levels of HBsAg in sera were measured at days 2 and 7 posthydrodynamic injection as shown in the upper and bottom panel, respectively. The results were presented as individual samples with mean ± SEM. Statistical significance was calculated by the Student's _t_-test and indicated by asterisks (*P < 0.05, **P < 0.01). (b) Analysis of intrahepatic HBV DNA by Southern blotting. 0.1 ng of pAAV/HBV 1.2 plasmid was loaded as a positive control. Two samples from each group were analyzed in this experiment. The band intensity was determined by software ImageJ. The numbers in the bottom indicate the relative intensities of the HBV expression plasmids in each sample. (c) Results of T7E1 assay. The percentage of mismatched sequences from two of the gRNA-P1- or gRNA-XCp-treated mice (Indel %) was determined by T7E1 assay. The numbers in the bottom indicate the percentage of Indel.
Similar articles
- Synthetic gRNA/Cas9 ribonucleoprotein targeting HBV DNA inhibits viral replication.
Zhang J, Zhang Y, Khanal S, Cao D, Zhao J, Dang X, Nguyen LNT, Schank M, Wu XY, Jiang Y, Ning S, Wang L, El Gazzar M, Moorman JP, Guo H, Yao ZQ. Zhang J, et al. J Med Virol. 2023 Jul;95(7):e28952. doi: 10.1002/jmv.28952. J Med Virol. 2023. PMID: 37455550 Free PMC article. - CRISPR/Cas9 System with Dual gRNAs Synergically Inhibit Hepatitis B Virus Replication.
Fei L, Sun S, Yang Q, Huang Y, Li Q, Tao S, Chen L. Fei L, et al. Discov Med. 2024 Jun;36(185):1169-1179. doi: 10.24976/Discov.Med.202436185.107. Discov Med. 2024. PMID: 38926103 - The potential and challenges of CRISPR-Cas in eradication of hepatitis B virus covalently closed circular DNA.
Yang HC, Chen PJ. Yang HC, et al. Virus Res. 2018 Jan 15;244:304-310. doi: 10.1016/j.virusres.2017.06.010. Epub 2017 Jun 13. Virus Res. 2018. PMID: 28627393 Review. - Dual gRNAs guided CRISPR/Cas9 system inhibits hepatitis B virus replication.
Wang J, Xu ZW, Liu S, Zhang RY, Ding SL, Xie XM, Long L, Chen XM, Zhuang H, Lu FM. Wang J, et al. World J Gastroenterol. 2015 Aug 28;21(32):9554-65. doi: 10.3748/wjg.v21.i32.9554. World J Gastroenterol. 2015. PMID: 26327763 Free PMC article. - In vivo Delivery Tools for Clustered Regularly Interspaced Short Palindromic Repeat/Associated Protein 9-Mediated Inhibition of Hepatitis B Virus Infection: An Update.
Kayesh MEH, Hashem MA, Kohara M, Tsukiyama-Kohara K. Kayesh MEH, et al. Front Microbiol. 2022 Jul 1;13:953218. doi: 10.3389/fmicb.2022.953218. eCollection 2022. Front Microbiol. 2022. PMID: 35847068 Free PMC article. Review.
Cited by
- The potential of HBV cure: an overview of CRISPR-mediated HBV gene disruption.
Yao ZQ, Schank MB, Zhao J, El Gazzar M, Wang L, Zhang Y, Hill AC, Banik P, Pyburn JS, Moorman JP. Yao ZQ, et al. Front Genome Ed. 2024 Oct 9;6:1467449. doi: 10.3389/fgeed.2024.1467449. eCollection 2024. Front Genome Ed. 2024. PMID: 39444780 Free PMC article. Review. - Tackling hepatitis B Virus with CRISPR/Cas9: advances, challenges, and delivery strategies.
Nair DM, Vajravelu LK, Thulukanam J, Paneerselvam V, Vimala PB, Lathakumari RH. Nair DM, et al. Virus Genes. 2024 Aug 28. doi: 10.1007/s11262-024-02105-3. Online ahead of print. Virus Genes. 2024. PMID: 39196289 Review. - Harnessing the evolving CRISPR/Cas9 for precision oncology.
Li T, Li S, Kang Y, Zhou J, Yi M. Li T, et al. J Transl Med. 2024 Aug 8;22(1):749. doi: 10.1186/s12967-024-05570-4. J Transl Med. 2024. PMID: 39118151 Free PMC article. Review. - Harnessing CRISPR technology for viral therapeutics and vaccines: from preclinical studies to clinical applications.
Zahedipour F, Zahedipour F, Zamani P, Jaafari MR, Sahebkar A. Zahedipour F, et al. Virus Res. 2024 Mar;341:199314. doi: 10.1016/j.virusres.2024.199314. Epub 2024 Jan 12. Virus Res. 2024. PMID: 38211734 Free PMC article. Review. - Novel Approaches to Inhibition of HBsAg Expression from cccDNA and Chromosomal Integrants: A Review.
Abdelwahed AH, Heineman BD, Wu GY. Abdelwahed AH, et al. J Clin Transl Hepatol. 2023 Dec 28;11(7):1485-1497. doi: 10.14218/JCTH.2023.00067. Epub 2023 Sep 19. J Clin Transl Hepatol. 2023. PMID: 38161502 Free PMC article. Review.
References
- Dandri M, Burda MR, Will H, Petersen J. Increased hepatocyte turnover and inhibition of woodchuck hepatitis B virus replication by adefovir in vitro do not lead to reduction of the closed circular DNA. Hepatology. 2000;32:139–146. - PubMed
- Tsiang M, Rooney JF, Toole JJ, Gibbs CS. Biphasic clearance kinetics of hepatitis B virus from patients during adefovir dipivoxil therapy. Hepatology. 1999;29:1863–1869. - PubMed
- Werle–Lapostolle B, Bowden S, Locarnini S, Wursthorn K, Petersen J, Lau G, et al. Persistence of cccDNA during the natural history of chronic hepatitis B and decline during adefovir dipivoxil therapy. Gastroenterology. 2004;126:1750–1758. - PubMed
- Caruntu FA, Molagic V. CccDNA persistence during natural evolution of chronic VHB infection. Rom J Gastroenterol. 2005;14:373–377. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials