Integrative structure-function mapping of the nucleoporin Nup133 suggests a conserved mechanism for membrane anchoring of the nuclear pore complex - PubMed (original) (raw)
. 2014 Nov;13(11):2911-26.
doi: 10.1074/mcp.M114.040915. Epub 2014 Aug 19.
Javier Fernandez-Martinez 2, Parthasarathy Sampathkumar 3, Anne Martel 4, Tsutomu Matsui 4, Hiro Tsuruta 4, Thomas M Weiss 4, Yi Shi 5, Ane Markina-Inarrairaegui 6, Jeffery B Bonanno 3, J Michael Sauder 7, Stephen K Burley 8, Brian T Chait 5, Steven C Almo 9, Michael P Rout 10, Andrej Sali 11
Affiliations
- PMID: 25139911
- PMCID: PMC4223481
- DOI: 10.1074/mcp.M114.040915
Integrative structure-function mapping of the nucleoporin Nup133 suggests a conserved mechanism for membrane anchoring of the nuclear pore complex
Seung Joong Kim et al. Mol Cell Proteomics. 2014 Nov.
Abstract
The nuclear pore complex (NPC) is the sole passageway for the transport of macromolecules across the nuclear envelope. Nup133, a major component in the essential Y-shaped Nup84 complex, is a large scaffold protein of the NPC's outer ring structure. Here, we describe an integrative modeling approach that produces atomic models for multiple states of Saccharomyces cerevisiae (Sc) Nup133, based on the crystal structures of the sequence segments and their homologs, including the related Vanderwaltozyma polyspora (Vp) Nup133 residues 55 to 502 (VpNup133(55-502)) determined in this study, small angle X-ray scattering profiles for 18 constructs of ScNup133 and one construct of VpNup133, and 23 negative-stain electron microscopy class averages of ScNup133(2-1157). Using our integrative approach, we then computed a multi-state structural model of the full-length ScNup133 and validated it with mutational studies and 45 chemical cross-links determined via mass spectrometry. Finally, the model of ScNup133 allowed us to annotate a potential ArfGAP1 lipid packing sensor (ALPS) motif in Sc and VpNup133 and discuss its potential significance in the context of the whole NPC; we suggest that ALPS motifs are scattered throughout the NPC's scaffold in all eukaryotes and play a major role in the assembly and membrane anchoring of the NPC in the nuclear envelope. Our results are consistent with a common evolutionary origin of Nup133 with membrane coating complexes (the protocoatomer hypothesis); the presence of the ALPS motifs in coatomer-like nucleoporins suggests an ancestral mechanism for membrane recognition present in early membrane coating complexes.
© 2014 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures
Fig. 1.
Integrative modeling approach for _Sc_Nup133. Our integrative modeling approach proceeds through three stages: (i) gathering of data; (ii) conformational sampling and scoring to produce a minimal ensemble of conformations consistent with SAXS profiles, EM class averages, template structures, and chemical cross-links; and (iii) analysis of the ensemble. The integrative modeling protocol was scripted in Python, based on our open-source IMP (Integrative Modeling Platform) package, release 2.2 (44).
Fig. 2.
Crystal structure of _Vp_Nup13355–502. A, overall fold of _Vp_Nup13355–502 β-propeller domain is shown in cartoon representation with color shading in blue to red from the N to the C terminus. Terminal residues of the disordered segments are marked with gray spheres. The location of the DA34-loop containing a potential ALPS motif is indicated by an arrow. Secondary structure elements are shown as defined by the DSSP (86) program. Strands within each propeller blade are marked in pink. B, comparison of the merged experimental SAXS profile (black) of _Vp_Nup13355–502 with the calculated SAXS profiles from the complete dimer model (χ = 12.4, red) and the complete monomer model (χ = 1.14, blue). The lower plot presents the residuals (calculated intensity/experimental intensity) of each calculated SAXS profile. The upper inset shows the SAXS profiles in the Guinier plot with an Rg fit of 24.4 ± 0.3 Å. The maximum particle size (_D_max) was 77.8 Å (determined experimentally). C, superposition of _Vp_Nup13355–502 (shown as a pink cartoon) and _Hs_Nup13375–477 (PDB: 1XKS; shown as a light-blue cartoon) (19) structures performed using the SSM (87) routine as implemented in COOT (34). D, electrostatic potential of _Vp_Nup13355–502 plotted onto its solvent accessible surface. Missing side chains and charges were assigned for the _Vp_Nup13355–502 structure on the PDB2PQR Web server (88), and the electrostatic surface was calculated using APBS (89) within PyMOL. Negative (−4 kT/e) and positive (+4 kT/e) potentials are shown in red and blue, respectively. The positively charged surface located adjacent to the DA34-loop (annotated as the potential ALPS motif) is marked by an ellipse in yellow. Phenylalanines at positions 275 and 276 are shown in sticks as a reference for the location of the DA34-loop.
Fig. 3.
Structure and dynamics of _Sc_Nup133 revealed through integrative modeling approach. A, the minimal ensemble of four conformations (the multi-state model), comprising a single major extended conformation with a population weight of 0.506 (blue) and three minor compact conformations with weights of 0.242 (red), 0.202 (cyan), and 0.050 (yellow), is shown. The most populated conformation (blue) was used as a reference for rigid body least-squares superposition of the remaining three conformations. The ab initio shape (represented as a gray envelope) computed from the experimental SAXS profile was also superposed for comparison. B, comparison of the merged experimental SAXS profile (black) of _Sc_Nup1332–1157 with the calculated SAXS profiles from the _Sc_Nup1332–1157 comparative model (χ = 6.27, red) and the ensemble of four conformations (χ = 1.54, blue). The lower plot presents the residuals (calculated intensity/experimental intensity) of each calculated SAXS profile. The upper inset shows the SAXS profiles in the Guinier plot with an Rg fit of 48.3 ± 0.6 Å. The maximum particle size (_D_max) was 169.2 Å (determined experimentally). C, the 23 negative-stain EM class averages are shown along with the projections of each of the four conformations. 22 EM class averages were assigned to at least one of the four conformations with high confidence, as highlighted in colored boxes.
Fig. 4.
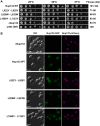
Mutational analysis of the _Sc_Nup133–_Sc_Nup84 interface. The double mutations at the predicted _Sc_Nup133–_Sc_Nup84 interface disrupted the association of _Sc_Nup133 with the rest of the _Sc_Nup84 complex. A, alignment of the Nup133 amino acid sequences of different organisms (indicated on the left) corresponding to the predicted Nup84 interaction surface in S. cerevisiae. The alignment was performed with ClustalW2 (90). The amino acids affected by the designed point mutations are indicated by an asterisk. B, affinity purification of nucleoporins associated with the Nup133-PrA mutants indicated above each gel line. The identity of each protein is indicated in blue letters on the right. Marker protein molecular weights are indicated in kDa on the left side. Immunoglobulin contaminants are identified in gray letters.
Fig. 5.
Phenotypic analysis of the _Sc_Nup133 point mutants. A, fitness analysis of the Nup133 mutants. 10-fold serial dilutions of strains expressing the Nup133-PrA construct indicated on the left side or carrying an empty plasmid (controls ΔNup133 and parental w303 strains) were spotted in minimal media plates without tryptophan and grown at the indicated temperatures for 2 to 3 days. A semi-quantitative score, in arbitrary units (AU), normalized to the wild-type fitness, is shown on the right. B, Nup133 mutants associated with the NPC. NPC association of the Nup133–GFP constructs indicated on the left side was analyzed via live-cell direct fluorescence microscopy in a ΔNup133, Nup170-mCherry mutant background. Cells were grown to early log phase at 25 °C. Differential interference contrast (DIC, left) and single-channel fluorescence (center and right columns) are shown. Scale bar = 5 mm.
Fig. 6.
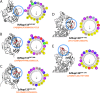
The potential ArfGAP1 lipid packing sensor (ALPS) motifs. Each of the potential ALPS motifs and sequences for _Hs_Nup133 (A), _Vp_Nup133 (B), _Sc_Nup133 (C), and _Sc_Nup120 (D, E) is presented. The potential ALPS motifs (red) were visualized in their corresponding structures (gray) using UCSF Chimera (37) and are highlighted by blue circles. The helical-wheel representations of the potential ALPS motifs are also shown, with an arrow in the center of the helical-wheel representing the direction and strength of the mean hydrophobic moment of the corresponding ALPS motif.
Similar articles
- Architecture of the linker-scaffold in the nuclear pore.
Petrovic S, Samanta D, Perriches T, Bley CJ, Thierbach K, Brown B, Nie S, Mobbs GW, Stevens TA, Liu X, Tomaleri GP, Schaus L, Hoelz A. Petrovic S, et al. Science. 2022 Jun 10;376(6598):eabm9798. doi: 10.1126/science.abm9798. Epub 2022 Jun 10. Science. 2022. PMID: 35679425 Free PMC article. - Structural and functional analysis of Nup120 suggests ring formation of the Nup84 complex.
Seo HS, Ma Y, Debler EW, Wacker D, Kutik S, Blobel G, Hoelz A. Seo HS, et al. Proc Natl Acad Sci U S A. 2009 Aug 25;106(34):14281-6. doi: 10.1073/pnas.0907453106. Epub 2009 Aug 11. Proc Natl Acad Sci U S A. 2009. PMID: 19706512 Free PMC article. - Structure, dynamics, evolution, and function of a major scaffold component in the nuclear pore complex.
Sampathkumar P, Kim SJ, Upla P, Rice WJ, Phillips J, Timney BL, Pieper U, Bonanno JB, Fernandez-Martinez J, Hakhverdyan Z, Ketaren NE, Matsui T, Weiss TM, Stokes DL, Sauder JM, Burley SK, Sali A, Rout MP, Almo SC. Sampathkumar P, et al. Structure. 2013 Apr 2;21(4):560-71. doi: 10.1016/j.str.2013.02.005. Epub 2013 Mar 14. Structure. 2013. PMID: 23499021 Free PMC article. - Membrane-coating lattice scaffolds in the nuclear pore and vesicle coats: commonalities, differences, challenges.
Leksa NC, Schwartz TU. Leksa NC, et al. Nucleus. 2010 Jul-Aug;1(4):314-8. doi: 10.4161/nucl.1.4.11798. Epub 2010 Mar 12. Nucleus. 2010. PMID: 21327078 Free PMC article. Review. - The structure of the nuclear pore complex.
Hoelz A, Debler EW, Blobel G. Hoelz A, et al. Annu Rev Biochem. 2011;80:613-43. doi: 10.1146/annurev-biochem-060109-151030. Annu Rev Biochem. 2011. PMID: 21495847 Review.
Cited by
- Dunking into the Lipid Bilayer: How Direct Membrane Binding of Nucleoporins Can Contribute to Nuclear Pore Complex Structure and Assembly.
Hamed M, Antonin W. Hamed M, et al. Cells. 2021 Dec 20;10(12):3601. doi: 10.3390/cells10123601. Cells. 2021. PMID: 34944108 Free PMC article. Review. - Development of a Prototype System for Archiving Integrative/Hybrid Structure Models of Biological Macromolecules.
Vallat B, Webb B, Westbrook JD, Sali A, Berman HM. Vallat B, et al. Structure. 2018 Jun 5;26(6):894-904.e2. doi: 10.1016/j.str.2018.03.011. Epub 2018 Apr 12. Structure. 2018. PMID: 29657133 Free PMC article. - High expression of nucleoporin 133 mRNA in bone marrow CD138+ cells is a poor prognostic factor in multiple myeloma.
Ibata S, Kobune M, Kikuchi S, Yoshida M, Miura S, Horiguchi H, Murase K, Iyama S, Takada K, Miyanishi K, Kato J. Ibata S, et al. Oncotarget. 2018 May 18;9(38):25127-25135. doi: 10.18632/oncotarget.25350. eCollection 2018 May 18. Oncotarget. 2018. PMID: 29861858 Free PMC article. - Principles for Integrative Structural Biology Studies.
Rout MP, Sali A. Rout MP, et al. Cell. 2019 May 30;177(6):1384-1403. doi: 10.1016/j.cell.2019.05.016. Cell. 2019. PMID: 31150619 Free PMC article. Review. - High Sensitivity Crosslink Detection Coupled With Integrative Structure Modeling in the Mass Spec Studio.
Sarpe V, Rafiei A, Hepburn M, Ostan N, Schryvers AB, Schriemer DC. Sarpe V, et al. Mol Cell Proteomics. 2016 Sep;15(9):3071-80. doi: 10.1074/mcp.O116.058685. Epub 2016 Jul 13. Mol Cell Proteomics. 2016. PMID: 27412762 Free PMC article.
References
- Grossman E., Medalia O., Zwerger M. (2012) Functional architecture of the nuclear pore complex. Annu. Rev. Biophys. 41, 557–584 - PubMed
- Alber F., Dokudovskaya S., Veenhoff L. M., Zhang W., Kipper J., Devos D., Suprapto A., Karni-Schmidt O., Williams R., Chait B. T., Sali A., Rout M. P. (2007) The molecular architecture of the nuclear pore complex. Nature 450, 695–701 - PubMed
- Alber F., Dokudovskaya S., Veenhoff L. M., Zhang W., Kipper J., Devos D., Suprapto A., Karni-Schmidt O., Williams R., Chait B. T., Rout M. P., Sali A. (2007) Determining the architectures of macromolecular assemblies. Nature 450, 683–694 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 GM083960/GM/NIGMS NIH HHS/United States
- P41 GM103314/GM/NIGMS NIH HHS/United States
- U54 GM074945/GM/NIGMS NIH HHS/United States
- P41 RR001209/RR/NCRR NIH HHS/United States
- U54 GM103511/GM/NIGMS NIH HHS/United States
- U54 GM094662/GM/NIGMS NIH HHS/United States
- P30 CA013330/CA/NCI NIH HHS/United States
- P41 GM103393/GM/NIGMS NIH HHS/United States
- U01 GM098256/GM/NIGMS NIH HHS/United States
- P41 GM109824/GM/NIGMS NIH HHS/United States
- R01 GM062427/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases