Cooperation between Epstein-Barr virus immune evasion proteins spreads protection from CD8+ T cell recognition across all three phases of the lytic cycle - PubMed (original) (raw)
Cooperation between Epstein-Barr virus immune evasion proteins spreads protection from CD8+ T cell recognition across all three phases of the lytic cycle
Laura L Quinn et al. PLoS Pathog. 2014.
Abstract
CD8+ T cell responses to Epstein-Barr virus (EBV) lytic cycle expressed antigens display a hierarchy of immunodominance, in which responses to epitopes of immediate-early (IE) and some early (E) antigens are more frequently observed than responses to epitopes of late (L) expressed antigens. It has been proposed that this hierarchy, which correlates with the phase-specific efficiency of antigen presentation, may be due to the influence of viral immune-evasion genes. At least three EBV-encoded genes, BNLF2a, BGLF5 and BILF1, have the potential to inhibit processing and presentation of CD8+ T cell epitopes. Here we examined the relative contribution of these genes to modulation of CD8+ T cell recognition of EBV lytic antigens expressed at different phases of the replication cycle in EBV-transformed B-cells (LCLs) which spontaneously reactivate lytic cycle. Selective shRNA-mediated knockdown of BNLF2a expression led to more efficient recognition of immediate-early (IE)- and early (E)-derived epitopes by CD8+ T cells, while knock down of BILF1 increased recognition of epitopes from E and late (L)-expressed antigens. Contrary to what might have been predicted from previous ectopic expression studies in EBV-negative model cell lines, the shRNA-mediated inhibition of BGLF5 expression in LCLs showed only modest, if any, increase in recognition of epitopes expressed in any phase of lytic cycle. These data indicate that whilst BNLF2a interferes with antigen presentation with diminishing efficiency as lytic cycle progresses (IE>E>>L), interference by BILF1 increases with progression through lytic cycle (IE<E<<L). Moreover, double-knockdown experiments showed that BILF1 and BNLF2a co-operate to further inhibit antigen presentation of L epitopes. Together, these data firstly indicate which potential immune-evasion functions are actually relevant in the context of lytic virus replication, and secondly identify lytic-cycle phase-specific effects that provide mechanistic insight into the immunodominance pattern seen for CD8+ T cell responses to EBV lytic antigens.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
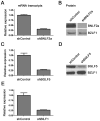
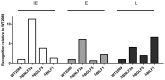
Figure 1. Knockdown of BNL2a, BILF1 and BGLF5 in transduced LCLs.
A) qRT-PCR was performed to measure the relative knockdown of BNLF2a transcript levels in shControl- and shBNLF2a-LCLs. BNLF2a-mRNA expression was normalized against BZLF1 and shown as relative BNLF2a expression. (B) BNLF2a protein knockdown was assessed using western blot analysis. Protein levels of BNLF2a and BZLF1 was measured in shControl- and shBNLF2a- LCLs. C) qRT-PCR assay of BGLF5 expression normalized against BZLF1 transcript level. Data are shown as BGLF5 expression relative to shControl LCLs. D) BGLF5 knockdown was confirmed at the protein level using western blot analysis. The expression of BGLF5 and BZLF1 protein was measured in shControl-LCLs and shBGLF5-LCLs. E) qRT-PCR assay of BILF1 expression normalized against BZLF1 transcript. Data are shown as BILF1 expression relative to that in shControl-LCLs.
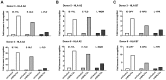
Figure 2. LCLs lacking in BNLF2a expression show increased presentation of epitopes derived from immediate early and early lytic antigens.
A) Donor 3 and 4 shBNLF2a-LCLs were used as targets for HLA-A2 restricted effector T cells specific to the YVL epitope of the IE gene BRLF1, the GLC epitope derived from an E gene BMLF1 and the FLD epitope which originates from the L expressed gene BALF4. Recognition was measured by ELISA for IFN- γ released by effector T cells. B) Donor 5 and 6 shBNLF2a-LCLs were used as targets for HLA-A2 restricted effector T cells specific to the YVL epitope of the IE gene BRLF1, the TLD epitope derived from an E gene BRLF1 and the WQW epitope which originates from the L expressed gene BNRF1. C) Donor 3 and 8 shBNLF2a-LCLs were used as targets for HLA-B7 restricted effector T cells specific to the DPY epitope of the IE gene BZLF1, the RPG epitope derived from an E gene BNLF2b and the YPR epitope which originates from the L expressed gene BNRF1. All representative data are shown as fold increase in recognition of shBNLF2a-LCLs compared to shControl transduced LCL counterparts, following normalisation of T cell recognition (IFN-γ release) against the expression levels of the antigen from which each epitope is derived.
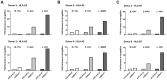
Figure 3. BGLF5 knockdown results in minimal increases in epitope recognition.
A) Relative recognition of donor 5 shBGLF5-LCLs, compared to shControl-LCLs, by a panel of HLA-A2 restricted CD8+ T cells specific for IE-YVL (BRLF1), E-GLC (BMLF1) and L-FLD (BALF4) epitopes. (B) Relative recognition of donor 6 shBGLF5-LCLs, compared to shControl-LCLs, by a panel of HLA-A2 restricted CD8+ T cells specific for IE-YVL (BRLF1), E-GLC (BMLF1) and L-WQW (BNRF1) epitopes. (C) Relative recognition of donor 3 HLA-B7 positive shBGLF5-LCLs, compared to Control LCLs, by HLA-B7 restricted T cells specific for the IE-DPY (BZLF1), E- RPG (BNLF2b) and L-YPR (BNRF1) epitopes. All representative data are shown as fold increase in recognition of shBGLF5-LCLs compared to shControl transduced LCL counterparts, following normalisation of T cell recognition (IFN-γ release) against the expression levels of the antigen from which each epitope is derived.
Figure 4. BILF1 predominantly interferes with peptide presentation to CD8+ T cells during late stage lytic cycle.
A) Donor 2 and 3 shBILF1-LCLs were used as targets for HLA-A2 restricted effector T cells specific to the YVL epitope of the IE gene BRLF1, the GLC epitope derived from an E gene BMLF1 and the FLD epitope which originates from the L expressed gene BALF4. B) Donor 5 and 6 shBILF1-LCLs were used as targets for HLA-A2 restricted effector T cells specific to the YVL epitope of the IE gene BRLF1, the TLD epitope derived from an E gene BMRF1 or the E-GLC epitope of BMLF1 and the WQW epitope which originates from the L expressed gene BNRF1. C) Donor 3 and 8 shBILF1-LCLs were used as targets for HLA-B7 restricted effector T cells specific to the DPY epitope of the IE gene BZLF1, the RPG epitope derived from an E gene BNLF2b and the YPR epitope which originates from the L expressed gene BNRF1. All representative data are shown as fold increase in recognition of shBILF1-LCLs compared to shControl transduced LCL counterparts, following normalisation of T cell recognition (IFN-γ release) against the expression levels of the antigen from which each epitope is derived.
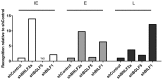
Figure 5. Direct comparison of the relative effects of BNLF2a, BGLF5 and BILF1 on T cell recognition of IE-YVL (BRLF1), E-GLC (BMLF1) and L-FLD (BALF4) epitopes.
Recognition of epitopes presented by each knockdown and control LCL was measured simultaneously. T cell recognition (IFN-γ release) was then normalised on the expression of each appropriate target mRNA transcript. Data are shown as recognition of knockdown LCLs relative to recognition of shControl LCLs. * For one target (IE-YVL in shBGLF5) expression of target transcripts was insufficient to assay, and no T cell recognition was observed, as indicated by ND.
Figure 6. Direct comparison of the relative effects of BNLF2a, BGLF5 and BILF1 on T cell recognition of IE, E and L lytic epitopes using B-cells transformed with ΔBNLF2a, ΔBGLF5 and ΔBILF1 viruses.
T cell recognition of epitopes presented by each LCL was measured simultaneously. Recognition (IFN-γ release) was normalised on the expression of each respective target mRNA transcript. Data is shown as recognition of knockout LCLs relative to recognition of WT-2089-LCLs and is the mean of two experiments using a total of two different IE-YVL (BRLF1) T cells, one E-GLC (BMLF1) and one E-TLD (BMRF1) T cell and two different L-FLD (BALF4) T cells. The complete set of individual results is presented in the Supplementary Information, Figures S6–S8.
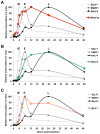
Figure 7. Expression kinetics of EBV lytic cycle.
EBV infected cells (Akata-BL) were synchronously induced into lytic cycle by ligation of the BCR. RNA was harvested at the indicated time points and cDNA was then synthesised followed by qRT-PCR analysis to detect the expression of IE-BZLF1, E-BMRF1 and L-BALF4 (A–C). The expression of these genes is compared to expression of BNLF2a (A), BGLF5 (B) and BILF1 (C). Samples were tested in duplicate and normalised to cellular GAPDH. Data are expressed as the relative number of transcripts as percentage of the maximum for each gene.
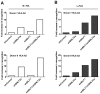
Figure 8. Relative recognition by IE- and L- specific, HLA-A2 restricted CD8+ T cell clones of LCLs lacking both BNLF2a and BILF1 expression.
A) Recognition of IE-YVL presented by donors 7 and 8 LCLs was measured simultaneously. T cell recognition (IFN-γ release) was normalised on the expression of BRLF1 mRNA transcript. Data are shown as recognition of single and double knockdown LCLs relative to shControl LCLs. (B) Recognition of L-FLD presented by each donor 7 and 8 LCLs was measured simultaneously. T cell recognition (IFN-γ release) was then normalised on the expression of BRLF1 mRNA transcript. Data are shown as recognition of single and double knockdown LCLs relative to recognition of shControl LCLs.
Figure 9. The relative roles of BNLF2a, BILF1 and BGLF5 in interfering with antigen presentation as lytic cycle progresses.
Diagram showing the strength of each immune evasion gene function at all stages of lytic cycle. BNLF2a is more potent at the IE time point and its effect diminishes as lytic cycle progresses. The potency of BILF1 increases as lytic cycle progresses. BGLF5 plays a minimal role throughout.
Similar articles
- Stage-specific inhibition of MHC class I presentation by the Epstein-Barr virus BNLF2a protein during virus lytic cycle.
Croft NP, Shannon-Lowe C, Bell AI, Horst D, Kremmer E, Ressing ME, Wiertz EJ, Middeldorp JM, Rowe M, Rickinson AB, Hislop AD. Croft NP, et al. PLoS Pathog. 2009 Jun;5(6):e1000490. doi: 10.1371/journal.ppat.1000490. Epub 2009 Jun 26. PLoS Pathog. 2009. PMID: 19557156 Free PMC article. - Epstein-Barr virus evasion of CD8(+) and CD4(+) T cell immunity via concerted actions of multiple gene products.
Ressing ME, Horst D, Griffin BD, Tellam J, Zuo J, Khanna R, Rowe M, Wiertz EJ. Ressing ME, et al. Semin Cancer Biol. 2008 Dec;18(6):397-408. doi: 10.1016/j.semcancer.2008.10.008. Epub 2008 Oct 25. Semin Cancer Biol. 2008. PMID: 18977445 Review. - CD8+ immunodominance among Epstein-Barr virus lytic cycle antigens directly reflects the efficiency of antigen presentation in lytically infected cells.
Pudney VA, Leese AM, Rickinson AB, Hislop AD. Pudney VA, et al. J Exp Med. 2005 Feb 7;201(3):349-60. doi: 10.1084/jem.20041542. Epub 2005 Jan 31. J Exp Med. 2005. PMID: 15684323 Free PMC article. - The Missing Link in Epstein-Barr Virus Immune Evasion: the BDLF3 Gene Induces Ubiquitination and Downregulation of Major Histocompatibility Complex Class I (MHC-I) and MHC-II.
Quinn LL, Williams LR, White C, Forrest C, Zuo J, Rowe M. Quinn LL, et al. J Virol. 2015 Oct 14;90(1):356-67. doi: 10.1128/JVI.02183-15. Print 2016 Jan 1. J Virol. 2015. PMID: 26468525 Free PMC article. - Immunodominance of lytic cycle antigens in Epstein-Barr virus-specific CD4+ T cell preparations for therapy.
Adhikary D, Behrends U, Boerschmann H, Pfünder A, Burdach S, Moosmann A, Witter K, Bornkamm GW, Mautner J. Adhikary D, et al. PLoS One. 2007 Jul 4;2(7):e583. doi: 10.1371/journal.pone.0000583. PLoS One. 2007. PMID: 17611619 Free PMC article. Review.
Cited by
- Cytokine Storm Syndromes Associated with Epstein-Barr Virus.
Verbist K, Nichols KE. Verbist K, et al. Adv Exp Med Biol. 2024;1448:227-248. doi: 10.1007/978-3-031-59815-9_16. Adv Exp Med Biol. 2024. PMID: 39117818 Review. - Epstein-Barr virus: the mastermind of immune chaos.
Silva JM, Alves CEC, Pontes GS. Silva JM, et al. Front Immunol. 2024 Feb 7;15:1297994. doi: 10.3389/fimmu.2024.1297994. eCollection 2024. Front Immunol. 2024. PMID: 38384471 Free PMC article. Review. - Evolution of functional antibodies following acute Epstein-Barr virus infection.
Karsten CB, Bartsch YC, Shin SA, Slein MD, Heller HM, Kolandaivelu K, Middeldorp JM, Alter G, Julg B. Karsten CB, et al. PLoS Pathog. 2022 Sep 6;18(9):e1010738. doi: 10.1371/journal.ppat.1010738. eCollection 2022 Sep. PLoS Pathog. 2022. PMID: 36067220 Free PMC article. - HLA Allele E*01:01 Is Associated with a Reduced Risk of EBV-Related Classical Hodgkin Lymphoma Independently of HLA-A*01/*02.
Martín P, Krsnik I, Navarro B, Provencio M, García JF, Bellas C, Vilches C, Gomez-Lozano N. Martín P, et al. PLoS One. 2015 Aug 11;10(8):e0135512. doi: 10.1371/journal.pone.0135512. eCollection 2015. PLoS One. 2015. PMID: 26261988 Free PMC article. - Functional Implications of Epstein-Barr Virus Lytic Genes in Carcinogenesis.
Yap LF, Wong AKC, Paterson IC, Young LS. Yap LF, et al. Cancers (Basel). 2022 Nov 24;14(23):5780. doi: 10.3390/cancers14235780. Cancers (Basel). 2022. PMID: 36497262 Free PMC article. Review.
References
- Zuckerman RA, Limaye AP (2013) Varicella zoster virus (VZV) and herpes simplex virus (HSV) in solid organ transplant patients. Am J Transplant 13 Suppl 3: 55–66; quiz 66. - PubMed
- Hebart H, Einsele H (2004) Clinical aspects of CMV infection after stem cell transplantation. Hum Immunol 65: 432–436. - PubMed
- Zerr DM, Corey L, Kim HW, Huang ML, Nguy L, et al. (2005) Clinical outcomes of human herpesvirus 6 reactivation after hematopoietic stem cell transplantation. Clin Infect Dis 40: 932–940. - PubMed
- Gottschalk S, Rooney CM, Heslop HE (2005) Post-transplant lymphoproliferative disorders. Annu Rev Med 56: 29–44. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- 15032/CRUK_/Cancer Research UK/United Kingdom
- MR/J002046/1/MRC_/Medical Research Council/United Kingdom
- G0901755/MRC_/Medical Research Council/United Kingdom
- C5575/A15032/CRUK_/Cancer Research UK/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials