Next-generation sequencing technologies: breaking the sound barrier of human genetics - PubMed (original) (raw)
Review
Next-generation sequencing technologies: breaking the sound barrier of human genetics
El Mustapha Bahassi et al. Mutagenesis. 2014 Sep.
Abstract
Demand for new technologies that deliver fast, inexpensive and accurate genome information has never been greater. This challenge has catalysed the rapid development of advances in next-generation sequencing (NGS). The generation of large volumes of sequence data and the speed of data acquisition are the primary advantages over previous, more standard methods. In 2013, the Food and Drug Administration granted marketing authorisation for the first high-throughput NG sequencer, Illumina's MiSeqDx, which allowed the development and use of a large number of new genome-based tests. Here, we present a review of template preparation, nucleic acid sequencing and imaging, genome assembly and alignment approaches as well as recent advances in current and near-term commercially available NGS instruments. We also outline the broad range of applications for NGS technologies and provide guidelines for platform selection to best address biological questions of interest. DNA sequencing has revolutionised biological and medical research, and is poised to have a similar impact on the practice of medicine. This tool is but one of an increasing arsenal of developing tools that enhance our capabilities to identify, quantify and functionally characterise the components of biological networks that keep us healthy or make us sick. Despite advances in other 'omic' technologies, DNA sequencing and analysis, in many respects, have played the leading role to date. The new technologies provide a bridge between genotype and phenotype, both in man and model organisms, and have revolutionised how risk of developing a complex human disease may be assessed. The generation of large DNA sequence data sets is producing a wealth of medically relevant information on a large number of individuals and populations that will potentially form the basis of truly individualised medical care in the future.
© The Author 2014. Published by Oxford University Press on behalf of the UK Environmental Mutagen Society. All rights reserved. For permissions, please e-mail: journals.permissions@oup.com.
Figures
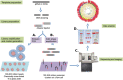
Fig. 1.
NGS technologies: template preparation, sequencing and imaging and data analysis. For WGS, gDNA is sheared by sonication or nebulisation to form fragments of 300–500 bp. Library amplification can be done by either emPCR or solid-phase amplification. In emPCR (A), a reaction mixture consisting of an oil–aqueous emulsion is created to encapsulate bead–DNA complexes into single aqueous droplets. PCR amplification is performed within these droplets to create beads containing several thousand copies of the same template sequence. EmPCR beads can be chemically attached to a glass slide or deposited into PicoTiterPlate wells. Solid-phase amplification (B) is composed of two basic steps: initial priming and extending of the single-stranded, single-molecule template, and bridge amplification of the immobilised template with immediately adjacent primers to form clusters. (C) Sequencing and imaging using one of the platforms described above. (D) Data analysis using the available software or an integrated workflow such as the GATK pipeline described below.
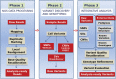
Fig. 2.
The GATK workflow for NGS data analysis.
Fig. 3.
Visualisation of NGS mutations in normal versus tumour tissue using IGV and Circos. (A) Detection of a SNP in a proneural brain tumour patient in_IDH1_ gene but not in normal DNA. A mutation C (blue) to T (red) is flagged in red in the tumour (top) but not in the normal (bottom). (B) Detection of CNVs in_EGFR_ gene in a classic GBM patient. The increase in copy number in the tumour is indicated by the large increase in the number of reads (top panel) compared with the low number of reads in the normal (bottom panel). (C) Detection of EGFR-vIII deletion in the tumour but not in the normal tissue of a GBM patient. The start and finish of the deletion are flagged in red in the tumour (bottom panel) but the deletion is absent in the normal tissue (upper panel). (D) A Circos diagram of a GBM patient. The outmost spine-like histogram (dark red) shows the coverage (10×) at 10 kb bin width. The numbered ring is the chromosomal ideogram, with each number indicating the position of an individual chromosome. The yellow grid ring shows the CNVs (at 10 kb bin width). The normal CNV values (defined by T1/C1 ratio) are represented by grey dots. Mono and bi-allelic deletions are represented by green and red circles, respectively. If the value of T1/C1 is >1.5 but <2.0, it is represented by a blue circle. Any T1/C1 value >2 is represented by a black circle. The CVN track clearly shows there is a chr7 trisomy. In addition, mono- and bi-allelic deletion at chr9 is also very prominent. The CNV pattern of chr10 indicates part of T1 tumour may have aneuploidy in chr10. The orange ring illustrates detected SNPs (8× coverage), validated by IGV analysis. The light yellow ring illustrates detected indels (8× coverage). Non-functional indels are represented by grey circles. Indels with functional impact (located in exon or UTR) are represented by red circles. The innermost circle shows various genomic rearrangements. Red, black, green and blue curves represent deletions (DEL), inversions (INV), inter- and intra-chromosomal translocation (CTX and ITX), respectively.
Similar articles
- Sequencing technologies - the next generation.
Metzker ML. Metzker ML. Nat Rev Genet. 2010 Jan;11(1):31-46. doi: 10.1038/nrg2626. Epub 2009 Dec 8. Nat Rev Genet. 2010. PMID: 19997069 Review. - [New generation sequencing in medical genetics].
Stoppa-Lyonnet D, Houdayer C. Stoppa-Lyonnet D, et al. Med Sci (Paris). 2012 Feb;28(2):123-4. doi: 10.1051/medsci/2012282001. Epub 2012 Feb 27. Med Sci (Paris). 2012. PMID: 22377291 French. No abstract available. - The next phase in human genetics.
Bansal V, Tewhey R, Topol EJ, Schork NJ. Bansal V, et al. Nat Biotechnol. 2011 Jan;29(1):38-9. doi: 10.1038/nbt.1757. Nat Biotechnol. 2011. PMID: 21221098 No abstract available. - DNA sequencing methods in human genetics and disease research.
Lehrach H. Lehrach H. F1000Prime Rep. 2013 Sep 2;5:34. doi: 10.12703/P5-34. F1000Prime Rep. 2013. PMID: 24049638 Free PMC article. Review. - Next-Generation Sequencing Technologies.
McCombie WR, McPherson JD, Mardis ER. McCombie WR, et al. Cold Spring Harb Perspect Med. 2019 Nov 1;9(11):a036798. doi: 10.1101/cshperspect.a036798. Cold Spring Harb Perspect Med. 2019. PMID: 30478097 Free PMC article. Review.
Cited by
- Targeted-Capture Next-Generation Sequencing in Diagnosis Approach of Pediatric Cholestasis.
Almes M, Spraul A, Ruiz M, Girard M, Roquelaure B, Laborde N, Gottrand F, Turquet A, Lamireau T, Dabadie A, Bonneton M, Thebaut A, Rohmer B, Lacaille F, Broué P, Fabre A, Mention-Mulliez K, Bouligand J, Jacquemin E, Gonzales E. Almes M, et al. Diagnostics (Basel). 2022 May 7;12(5):1169. doi: 10.3390/diagnostics12051169. Diagnostics (Basel). 2022. PMID: 35626323 Free PMC article. - Hobnail variant of papillary thyroid carcinoma: molecular profiling and comparison to classical papillary thyroid carcinoma, poorly differentiated thyroid carcinoma and anaplastic thyroid carcinoma.
Teng L, Deng W, Lu J, Zhang J, Ren X, Duan H, Chuai S, Duan F, Gao W, Lu T, Wu H, Liang Z. Teng L, et al. Oncotarget. 2017 Mar 28;8(13):22023-22033. doi: 10.18632/oncotarget.15786. Oncotarget. 2017. PMID: 28423545 Free PMC article. - Impact of germline and somatic missense variations on drug binding sites.
Yan C, Pattabiraman N, Goecks J, Lam P, Nayak A, Pan Y, Torcivia-Rodriguez J, Voskanian A, Wan Q, Mazumder R. Yan C, et al. Pharmacogenomics J. 2017 Mar;17(2):128-136. doi: 10.1038/tpj.2015.97. Epub 2016 Jan 26. Pharmacogenomics J. 2017. PMID: 26810135 Free PMC article. - Software updates in the Illumina HiSeq platform affect whole-genome bisulfite sequencing.
Toh H, Shirane K, Miura F, Kubo N, Ichiyanagi K, Hayashi K, Saitou M, Suyama M, Ito T, Sasaki H. Toh H, et al. BMC Genomics. 2017 Jan 5;18(1):31. doi: 10.1186/s12864-016-3392-9. BMC Genomics. 2017. PMID: 28056787 Free PMC article.
References
- Hayden E. C. (2014)Is the $1,000 genome for real? Nature News, January15.
- Robertson J. A. (2003)The $1000 genome: ethical and legal issues in whole genome sequencing of individuals. Am. J. Bioeth., 3,W-IF1. - PubMed
- Green E. D., Guyer M. S. (2011)Charting a course for genomic medicine from base pairs to bedside. Nature, 470,204–213. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources