Comparative analysis of pseudogenes across three phyla - PubMed (original) (raw)
Comparative Study
. 2014 Sep 16;111(37):13361-6.
doi: 10.1073/pnas.1407293111. Epub 2014 Aug 25.
Baikang Pei 2, Jing Leng 2, Adam Frankish 3, Yan Zhang 2, Suganthi Balasubramanian 4, Rachel Harte 5, Daifeng Wang 2, Michael Rutenberg-Schoenberg 2, Wyatt Clark 2, Mark Diekhans 5, Joel Rozowsky 4, Tim Hubbard 3, Jennifer Harrow 3, Mark B Gerstein 6
Affiliations
- PMID: 25157146
- PMCID: PMC4169933
- DOI: 10.1073/pnas.1407293111
Comparative Study
Comparative analysis of pseudogenes across three phyla
Cristina Sisu et al. Proc Natl Acad Sci U S A. 2014.
Abstract
Pseudogenes are degraded fossil copies of genes. Here, we report a comparison of pseudogenes spanning three phyla, leveraging the completed annotations of the human, worm, and fly genomes, which we make available as an online resource. We find that pseudogenes are lineage specific, much more so than protein-coding genes, reflecting the different remodeling processes marking each organism's genome evolution. The majority of human pseudogenes are processed, resulting from a retrotranspositional burst at the dawn of the primate lineage. This burst can be seen in the largely uniform distribution of pseudogenes across the genome, their preservation in areas with low recombination rates, and their preponderance in highly expressed gene families. In contrast, worm and fly pseudogenes tell a story of numerous duplication events. In worm, these duplications have been preserved through selective sweeps, so we see a large number of pseudogenes associated with highly duplicated families such as chemoreceptors. However, in fly, the large effective population size and high deletion rate resulted in a depletion of the pseudogene complement. Despite large variations between these species, we also find notable similarities. Overall, we identify a broad spectrum of biochemical activity for pseudogenes, with the majority in each organism exhibiting varying degrees of partial activity. In particular, we identify a consistent amount of transcription (∼15%) across all species, suggesting a uniform degradation process. Also, we see a uniform decay of pseudogene promoter activity relative to their coding counterparts and identify a number of pseudogenes with conserved upstream sequences and activity, hinting at potential regulatory roles.
Keywords: functional genomics; genome annotation; transcriptomics.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
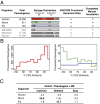
Fig. 1.
Annotation, classification, and evolution. (A) Pseudogene annotation and ENCODE functional data availability. (B) Distribution of processed pseudogenes as a function of pseudogene age (sequence similarity to parent genes) for human (Left) and worm and fly (Right). (C) Pseudogene disablement variation and density.
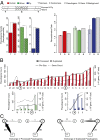
Fig. 2.
Localization and mobility. (A, Left) The relative chromosomal localization preference for pseudogenes in human, worm, and fly. (Right) Average recombination rates for pseudogenes, protein-coding genes, and genomic background. (B) Distributions of processed and duplicated pseudogenes across chromosomes, sorted by length. (C) Pseudogene exchange between sex chromosomes and autosomes in humans.
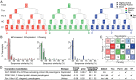
Fig. 3.
Orthologs, paralogs, and families. (A) Venn diagrams showing the total number of orthologous genes and pseudogenes, in human, worm, and fly. (Right) Pseudogene orthologs between human and mouse. (B) Per chromosome distribution of RpS6 pseudogenes in human, worm, and fly. (C) Comparative distribution of pseudogene and paralogs per gene. (D) Top pseudogene families that give rise to 25% of the total number of pseudogenes in each organism (Left, family type; Right, number of pseudogenes). Oval rows indicate the collapse of two or more consecutive families of the same type. 7tm, G protein-coupled receptors; His, histone; IG, Ig; Kin, kinase; Ploop, P-loop NTPase proteins; Ribo, ribosomal proteins; RRM, RNA recognition motifs; Struct, structural protein; ZnF, Zinc finger proteins (TF); Ubq, ubiquitination proteins; Motor, kinesin motor domain proteins; SAP, SAP domain proteins.
Fig. 4.
Pseudogene activity. (A) Distribution of pseudogenes as a function of various activity features: transcription (Tnx), active chromatin (AC), and presence of active Pol II and TF binding sites in the upstream region. (B) Conservation of the upstream sequences in processed and duplicated pseudogenes compared with paralogs. (C) Conservation of an upstream sequence activity mark (H3K27Ac) in pseudogene-parent pairs vs. parent-paralogs. +, active H3K27Ac; −, inactivity. We find that the majority of parent–paralog pairs have coordinated H3K27Ac activity (larger diagonal values) as opposed to parent–pseudogene pairs (larger off-diagonal values). (D) Functional pseudogene candidates with translation evidence.
Similar articles
- Digging for dead genes: an analysis of the characteristics of the pseudogene population in the Caenorhabditis elegans genome.
Harrison PM, Echols N, Gerstein MB. Harrison PM, et al. Nucleic Acids Res. 2001 Feb 1;29(3):818-30. doi: 10.1093/nar/29.3.818. Nucleic Acids Res. 2001. PMID: 11160906 Free PMC article. - Identification of pseudogenes in the Drosophila melanogaster genome.
Harrison PM, Milburn D, Zhang Z, Bertone P, Gerstein M. Harrison PM, et al. Nucleic Acids Res. 2003 Feb 1;31(3):1033-7. doi: 10.1093/nar/gkg169. Nucleic Acids Res. 2003. PMID: 12560500 Free PMC article. - Comprehensive analysis of amino acid and nucleotide composition in eukaryotic genomes, comparing genes and pseudogenes.
Echols N, Harrison P, Balasubramanian S, Luscombe NM, Bertone P, Zhang Z, Gerstein M. Echols N, et al. Nucleic Acids Res. 2002 Jun 1;30(11):2515-23. doi: 10.1093/nar/30.11.2515. Nucleic Acids Res. 2002. PMID: 12034841 Free PMC article. - Studying genomes through the aeons: protein families, pseudogenes and proteome evolution.
Harrison PM, Gerstein M. Harrison PM, et al. J Mol Biol. 2002 May 17;318(5):1155-74. doi: 10.1016/s0022-2836(02)00109-2. J Mol Biol. 2002. PMID: 12083509 Review. - Computational Methods for Pseudogene Annotation Based on Sequence Homology.
Harrison PM. Harrison PM. Methods Mol Biol. 2021;2324:35-48. doi: 10.1007/978-1-0716-1503-4_3. Methods Mol Biol. 2021. PMID: 34165707 Review.
Cited by
- Human-specific features and developmental dynamics of the brain N-glycome.
Klarić TS, Gudelj I, Santpere G, Novokmet M, Vučković F, Ma S, Doll HM, Risgaard R, Bathla S, Karger A, Nairn AC, Luria V, Bečeheli I, Sherwood CC, Ely JJ, Hof PR, Sousa AMM, Josić D, Lauc G, Sestan N. Klarić TS, et al. Sci Adv. 2023 Dec 8;9(49):eadg2615. doi: 10.1126/sciadv.adg2615. Epub 2023 Dec 6. Sci Adv. 2023. PMID: 38055821 Free PMC article. - A gene graveyard in the genome of the fungus Podospora comata.
Silar P, Dauget JM, Gautier V, Grognet P, Chablat M, Hermann-Le Denmat S, Couloux A, Wincker P, Debuchy R. Silar P, et al. Mol Genet Genomics. 2019 Feb;294(1):177-190. doi: 10.1007/s00438-018-1497-3. Epub 2018 Oct 4. Mol Genet Genomics. 2019. PMID: 30288581 - Primate piRNA Cluster Evolution Suggests Limited Relevance of Pseudogenes in piRNA-Mediated Gene Regulation.
Gebert D, Zischler H, Rosenkranz D. Gebert D, et al. Genome Biol Evol. 2019 Apr 1;11(4):1088-1104. doi: 10.1093/gbe/evz060. Genome Biol Evol. 2019. PMID: 30888404 Free PMC article. - Evolutionary Origins of Pseudogenes and Their Association with Regulatory Sequences in Plants.
Xie J, Li Y, Liu X, Zhao Y, Li B, Ingvarsson PK, Zhang D. Xie J, et al. Plant Cell. 2019 Mar;31(3):563-578. doi: 10.1105/tpc.18.00601. Epub 2019 Feb 13. Plant Cell. 2019. PMID: 30760562 Free PMC article. - GENCODE Pseudogenes.
Sisu C. Sisu C. Methods Mol Biol. 2021;2324:67-82. doi: 10.1007/978-1-0716-1503-4_5. Methods Mol Biol. 2021. PMID: 34165709
References
- Zhang Z, et al. PseudoPipe: An automated pseudogene identification pipeline. Bioinformatics. 2006;22(12):1437–1439. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases