ASTRAL: genome-scale coalescent-based species tree estimation - PubMed (original) (raw)
ASTRAL: genome-scale coalescent-based species tree estimation
S Mirarab et al. Bioinformatics. 2014.
Abstract
Motivation: Species trees provide insight into basic biology, including the mechanisms of evolution and how it modifies biomolecular function and structure, biodiversity and co-evolution between genes and species. Yet, gene trees often differ from species trees, creating challenges to species tree estimation. One of the most frequent causes for conflicting topologies between gene trees and species trees is incomplete lineage sorting (ILS), which is modelled by the multi-species coalescent. While many methods have been developed to estimate species trees from multiple genes, some which have statistical guarantees under the multi-species coalescent model, existing methods are too computationally intensive for use with genome-scale analyses or have been shown to have poor accuracy under some realistic conditions.
Results: We present ASTRAL, a fast method for estimating species trees from multiple genes. ASTRAL is statistically consistent, can run on datasets with thousands of genes and has outstanding accuracy-improving on MP-EST and the population tree from BUCKy, two statistically consistent leading coalescent-based methods. ASTRAL is often more accurate than concatenation using maximum likelihood, except when ILS levels are low or there are too few gene trees.
Availability and implementation: ASTRAL is available in open source form at https://github.com/smirarab/ASTRAL/. Datasets studied in this article are available at http://www.cs.utexas.edu/users/phylo/datasets/astral.
Supplementary information: Supplementary data are available at Bioinformatics online.
© The Author 2014. Published by Oxford University Press.
Figures
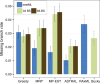
Fig. 1.
Species tree estimation error on the default mammalian datasets with 37 genes and 400 genes (half with 500 bp and half with 1000 bp and with 71% mean BS). We show the missing branch rates for estimated species trees computed using summary methods (MRP, MP-EST, greedy, BUCKy-pop and ASTRAL) as well as concatenation using RAxML. Results are shown for running summary methods on maximum likelihood gene trees (bestML) and on the set of all bootstrap replicates from all genes (All BS), as well as the greedy consensus of running summary methods on individual bootstrap replicates from all genes (MLBS). CA-ML is run on the true alignment. Average and standard error shown based on 20 replicates
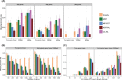
Fig. 2.
Species tree estimation error on the simulated mammalian datasets. We show the missing branch rates for estimated species trees computed using summary methods (MRP, MP-EST, greedy and ASTRAL) as well as CA-ML. Summary methods are run on RAxML bestML gene trees. We also show performance of summary methods on the true gene trees. Subfigure (A) shows results under default levels of ILS, varying the number of genes and gene tree resolution; (B) shows results under increased ILS levels, varying the number of genes, and on both true gene trees and estimated gene trees and (C) shows results on 200 genes, varying the amount of ILS from very low (5× species tree branch lengths) to very high (0.2× species tree branch lengths)
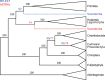
Fig. 3.
Analysis of the Song et al. mammals dataset using ASTRAL and MP-EST. We show the result of applying ASTRAL and MP-EST to 424 gene trees on 37-taxon mammalian species. MP-EST is based on rooted gene trees; ASTRAL is based on unrooted gene trees, and then rooted at the branch leading to the outgroup. Branch support values in black are for both methods, those in red are for ASTRAL and values in blue are for MP-EST. See
Supplementary Materials
for trees with full resolution
Similar articles
- ASTRAL-II: coalescent-based species tree estimation with many hundreds of taxa and thousands of genes.
Mirarab S, Warnow T. Mirarab S, et al. Bioinformatics. 2015 Jun 15;31(12):i44-52. doi: 10.1093/bioinformatics/btv234. Bioinformatics. 2015. PMID: 26072508 Free PMC article. - A comparative study of SVDquartets and other coalescent-based species tree estimation methods.
Chou J, Gupta A, Yaduvanshi S, Davidson R, Nute M, Mirarab S, Warnow T. Chou J, et al. BMC Genomics. 2015;16 Suppl 10(Suppl 10):S2. doi: 10.1186/1471-2164-16-S10-S2. Epub 2015 Oct 2. BMC Genomics. 2015. PMID: 26449249 Free PMC article. - ASTRID: Accurate Species TRees from Internode Distances.
Vachaspati P, Warnow T. Vachaspati P, et al. BMC Genomics. 2015;16 Suppl 10(Suppl 10):S3. doi: 10.1186/1471-2164-16-S10-S3. Epub 2015 Oct 2. BMC Genomics. 2015. PMID: 26449326 Free PMC article. - Challenges in Species Tree Estimation Under the Multispecies Coalescent Model.
Xu B, Yang Z. Xu B, et al. Genetics. 2016 Dec;204(4):1353-1368. doi: 10.1534/genetics.116.190173. Genetics. 2016. PMID: 27927902 Free PMC article. Review. - Estimating phylogenetic trees from genome-scale data.
Liu L, Xi Z, Wu S, Davis CC, Edwards SV. Liu L, et al. Ann N Y Acad Sci. 2015 Dec;1360:36-53. doi: 10.1111/nyas.12747. Epub 2015 Apr 14. Ann N Y Acad Sci. 2015. PMID: 25873435 Review.
Cited by
- Machine learning based imputation techniques for estimating phylogenetic trees from incomplete distance matrices.
Bhattacharjee A, Bayzid MS. Bhattacharjee A, et al. BMC Genomics. 2020 Jul 20;21(1):497. doi: 10.1186/s12864-020-06892-5. BMC Genomics. 2020. PMID: 32689946 Free PMC article. - Draft genome of the famous ornamental plant Paeonia suffruticosa.
Lv S, Cheng S, Wang Z, Li S, Jin X, Lan L, Yang B, Yu K, Ni X, Li N, Hou X, Huang G, Wang J, Dong Y, Wang E, Huang J, Zhang G, Zhang C. Lv S, et al. Ecol Evol. 2020 May 12;10(11):4518-4530. doi: 10.1002/ece3.5965. eCollection 2020 Jun. Ecol Evol. 2020. PMID: 32551041 Free PMC article. - A story from the Miocene: Clock-dated phylogeny of Sisymbrium L. (Sisymbrieae, Brassicaceae).
Žerdoner Čalasan A, German DA, Hurka H, Neuffer B. Žerdoner Čalasan A, et al. Ecol Evol. 2021 Mar 2;11(6):2573-2595. doi: 10.1002/ece3.7217. eCollection 2021 Mar. Ecol Evol. 2021. PMID: 33767822 Free PMC article. - Genome report: chromosome-level draft assemblies of the snow leopard, African leopard, and tiger (Panthera uncia, Panthera pardus pardus, and Panthera tigris).
Armstrong EE, Campana MG, Solari KA, Morgan SR, Ryder OA, Naude VN, Samelius G, Sharma K, Hadly EA, Petrov DA. Armstrong EE, et al. G3 (Bethesda). 2022 Dec 1;12(12):jkac277. doi: 10.1093/g3journal/jkac277. G3 (Bethesda). 2022. PMID: 36250809 Free PMC article. - Multi-tissue transcriptomes of caecilian amphibians highlight incomplete knowledge of vertebrate gene families.
Torres-Sánchez M, Creevey CJ, Kornobis E, Gower DJ, Wilkinson M, San Mauro D. Torres-Sánchez M, et al. DNA Res. 2019 Feb 1;26(1):13-20. doi: 10.1093/dnares/dsy034. DNA Res. 2019. PMID: 30351380 Free PMC article.
References
- Allman ES, et al. Identifying the rooted species tree from the distribution of unrooted gene trees under the coalescent. J. Math. Biol. 2011;62:833–862. - PubMed
- Bayzid MS, Warnow T. Naive binning improves phylogenomic analyses. Bioinformatics. 2013;29:2277–2284. - PubMed
- DeGiorgio M, Degnan JH. Robustness to divergence time underestimation when inferring species trees from estimated gene trees. Syst. Biol. 2014;63:66–82. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials