Data use under the NIH GWAS data sharing policy and future directions - PubMed (original) (raw)
Laura Lyman Rodriguez, Michael Feolo, Elizabeth Gillanders, Erin M Ramos, Joni L Rutter, Stephen Sherry, Vivian Ota Wang, Alice Bailey, Rebecca Baker, Mark Caulder, Emily L Harris, Kristofor Langlais, Hilary Leeds, Erin Luetkemeier, Taunton Paine, Tamar Roomian, Kimberly Tryka, Amy Patterson, Eric D Green; National Institutes of Health Genomic Data Sharing Governance Committees
- PMID: 25162809
- PMCID: PMC4182942
- DOI: 10.1038/ng.3062
Data use under the NIH GWAS data sharing policy and future directions
Dina N Paltoo et al. Nat Genet. 2014 Sep.
Abstract
In 2007, the US National Institutes of Health (NIH) introduced the Genome-Wide Association Studies (GWAS) Policy and the database of Genotypes and Phenotypes (dbGaP) to facilitate 'controlled' access to GWAS data based on participants' informed consent. dbGaP has provided 2,221 investigators access to 304 studies, resulting in 924 publications and significant scientific advances. Following on this success, the 2014 Genomic Data Sharing Policy will extend the GWAS Policy to additional data types.
Supplementary information: The online version of this article (doi:10.1038/ng.3062) contains supplementary material, which is available to authorized users.
Conflict of interest statement
The author declare no competing financial interests.
Figures
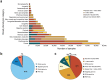
Figure 1. Number of dbGaP samples by broad phenotypic category and genomic technology.
(a) GWAS samples in dbGaP by broad phenotypic category and size of SNP array. Original submitting investigators select a broad phenotypic category for their studies from standard National Library of Medicine Medical Subject Headings (MeSH) at the time of study registration. The phenotypic categories of GWAS samples are shown by the scale of genotypic array used in the study (n = 393,729). (b) dbGaP samples by type of next-generation DNA sequencing performed. In addition to GWAS data, dbGaP maintains data collected using next-generation sequencing technologies, including sequencing of all genomic DNA (whole-genome), some genomic DNA (targeted genome), DNA expressed as RNA (whole-exome) and RNA-seq studies (n = 65,770). (c) dbGaP samples by type of genomic analysis performed (n = 459,499).
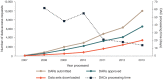
Figure 2. dbGaP data access activity from April 2007 to 1 December 2013.
Shown is the number of DARs submitted to the NIH, approved DARs, downloaded data sets and average time for DACs to process DARs.
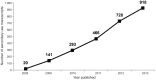
Figure 3. Publications describing research involving the secondary analyses of dbGaP data.
The cumulative number of publications reported by approved users of dbGaP data was collected from annual reports from 2007 to 2013.
Similar articles
- The genome-wide association studies data sharing policy.
Shurin SB. Shurin SB. Clin Transl Sci. 2008 Sep;1(2):91. doi: 10.1111/j.1752-8062.2008.00044.x. Clin Transl Sci. 2008. PMID: 20443825 Free PMC article. No abstract available. - Biospecimen policy: Family matters.
Hudson KL, Collins FS. Hudson KL, et al. Nature. 2013 Aug 8;500(7461):141-2. doi: 10.1038/500141a. Nature. 2013. PMID: 23925224 Free PMC article. - NIH broadens genomic data-sharing policy.
[No authors listed] [No authors listed] Cancer Discov. 2014 Dec;4(12):OF4. doi: 10.1158/2159-8290.CD-NB2014-148. Epub 2014 Oct 9. Cancer Discov. 2014. PMID: 25477121 - NIH's genomic data sharing policy: timing and tradeoffs.
Contreras JL. Contreras JL. Trends Genet. 2015 Feb;31(2):55-7. doi: 10.1016/j.tig.2014.12.006. Epub 2015 Jan 22. Trends Genet. 2015. PMID: 25620797 Review. - South Korea: in the midst of a privacy reform centered on data sharing.
Kim H, Kim SY, Joly Y. Kim H, et al. Hum Genet. 2018 Aug;137(8):627-635. doi: 10.1007/s00439-018-1920-1. Epub 2018 Aug 18. Hum Genet. 2018. PMID: 30121900 Free PMC article. Review.
Cited by
- Toward Better Semantic Interoperability of Data Element Repositories in Medicine: Analysis Study.
Hu Z, Wang A, Duan Y, Zhou J, Hu W, Wu S. Hu Z, et al. JMIR Med Inform. 2024 Sep 30;12:e60293. doi: 10.2196/60293. JMIR Med Inform. 2024. PMID: 39348178 Free PMC article. - When are your cells no longer your own?
[No authors listed] [No authors listed] Nat Biotechnol. 2023 Sep;41(9):1177. doi: 10.1038/s41587-023-01968-5. Nat Biotechnol. 2023. PMID: 37699988 No abstract available. - An agenda-setting paper on data sharing platforms: euCanSHare workshop.
Devriendt T, Ammann C, W Asselbergs F, Bernier A, Costas R, Friedrich MG, Gelpi JL, Jarvelin MR, Kuulasmaa K, Lekadir K, Mayrhofer MT, Papez V, Pasterkamp G, Petersen SE, Schmidt CO, Schulz-Menger J, Söderberg S, Shabani M, Veronesi G, Viezzer DS, Borry P. Devriendt T, et al. Open Res Eur. 2021 Nov 23;1:80. doi: 10.12688/openreseurope.13860.2. eCollection 2021. Open Res Eur. 2021. PMID: 37645200 Free PMC article. - Empirical validation of an automated approach to data use oversight.
Cabili MN, Lawson J, Saltzman A, Rushton G, O'Rourke P, Wilbanks J, Rodriguez LL, Nyronen T, Courtot M, Donnelly S, Philippakis AA. Cabili MN, et al. Cell Genom. 2021 Nov 10;1(2):100031. doi: 10.1016/j.xgen.2021.100031. eCollection 2021 Nov 10. Cell Genom. 2021. PMID: 36778584 Free PMC article. - Identifying Datasets for Cross-Study Analysis in dbGaP using PhenX.
Pan H, Bakalov V, Cox L, Engle ML, Erickson SW, Feolo M, Guo Y, Huggins W, Hwang S, Kimura M, Krzyzanowski M, Levy J, Phillips M, Qin Y, Williams D, Ramos EM, Hamilton CM. Pan H, et al. Sci Data. 2022 Sep 1;9(1):532. doi: 10.1038/s41597-022-01660-4. Sci Data. 2022. PMID: 36050327 Free PMC article.
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources