Optimization of genome engineering approaches with the CRISPR/Cas9 system - PubMed (original) (raw)
Optimization of genome engineering approaches with the CRISPR/Cas9 system
Kai Li et al. PLoS One. 2014.
Abstract
Designer nucleases such as TALENS and Cas9 have opened new opportunities to scarlessly edit the mammalian genome. Here we explored several parameters that influence Cas9-mediated scarless genome editing efficiency in murine embryonic stem cells. Optimization of transfection conditions and enriching for transfected cells are critical for efficiently recovering modified clones. Paired gRNAs and wild-type Cas9 efficiently create programmed deletions, which facilitate identification of targeted clones, while paired gRNAs and the Cas9D10A nickase generated smaller targeted indels with lower chance of off-target mutagenesis. Genome editing is also useful for programmed introduction of exogenous DNA sequences at a target locus. Increasing the length of the homology arms of the homology-directed repair template strongly enhanced targeting efficiency, while increasing the length of the DNA insert reduced it. Together our data provide guidance on optimal design of scarless gene knockout, modification, or knock-in experiments using Cas9 nuclease.
Conflict of interest statement
Competing Interests: The authors have declared that no competing interests exist.
Figures
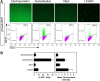
Figure 1. Optimization of mESC transfection.
A. pCas9-GFP expression plasmid was transfected into mESCs by the indicated method. GFP fluorescence was assessed by fluorescent microscopy and FACS. Numbers indicate the fraction of GFP-expressing cells. B. Percent of transfected cells and mean fluorescence intensity of GFP+ cells. n = 3. Graphs show mean ± s.e.m.
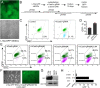
Figure 2. Cas9 gene knockout using single or dual gRNAs.
A. mESC cell line L10a-GFP, in which one Rosa26 locus expresses GFP. B. Experimental outline and gRNAs. gRNAs #9 and #5 generate 5′ overhangs for dual nickase strategy. C. Single gRNA-directed GFP inactivation. D. Comparison of gRNA alone or gRNA+HDR donor containing a translational stop signal. n = 3. Bar = s.e.m. Red circles indicate individual data points. E. Gene inactivation frequency of single compared to paired gRNAs. F–G. Assessment of GFP expression in mESCs in the GFP-FACS fraction by fluorescent microscopy and western blotting. H. Effective gene knockout using dual gRNAs and the hCas9-D10A nickase mutant.
Figure 3. Knockout of 3 congenital heart disease genes in mESCs.
A. Experimental outline. Paired gRNAs are designed to induce a deletion of about 100 bp. B. PCR genotyping of pooled ESC genomic DNA after Smad2 targeting. The mutant PCR product predominated. C. Surveyor nuclease on pooled ESC genomic DNA. Red arrowheads indicate nuclease cleavage products indicative of heterodimers. D. PCR genotyping of individual clones showing Smad2Δ/Δ, Smad2+/+, and Smad2+/Δ clones. E–F. Frequency of Smad2 genotypes amongst clonal outgrowths, after enrichment for transfected cells by GFP FACS or by transient puromycin selection. G. Western blot measurement of Smad2 expression in wild-type and Smad2Δ/Δ clones. H. Frequency of Mll2 genotypes amongst 39 genotyped clones. I. Confirmation of Mll2 inactivation in individual Mll2Δ/Δ clones by qRTPCR. J. Frequency of Chd7 genotypes amongst 48 genotyped clones. K. Confirmation of Chd7 inactivation in individual Chd7Δ/Δ clones by qRTPCR.
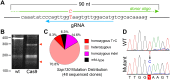
Figure 4. Cas9-mediated introduction of a single base change.
A. Sap130 targeting strategy. The blue arrow indicates the gRNA targeting the reverse strand with the tail and tip representing the 5′ and 3′ ends, respectively. The HDR donor oligo is indicated by the green arrows. The T>C change is indicated in red. B. Surveyor nuclease assay on pooled ESC genomic DNA. Red arrowheads indicate nuclease cleavage products, diagnostic of heterodimers. C. Distribution of Sap130 mutations. D. Sequence chromatograms showing homozygous T>C mutation.
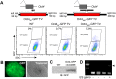
Figure 5. Cas9-mediated targeted knockin.
A. Mef2c traditional targeting vector containing long homology arms, fl-bio epitope tag, and positive selection cassette. Efficiency of traditional and Cas9-mediated targeting strategies is shown. B. Cas9-mediated targeting using a 50 bp homology arms and no selectable marker. C. PCR genotyping of pooled ESC genomic DNA. Upper band is diagnostic of knockin. D. Confirmation of knockin and effective epitope tagging by streptavidin pulldown of in vivo biotinylated Mef2c. E. Cas9-mediated knockin of a longer insert. F. PCR genotyping of pooled ESC genomic DNA. The band is diagnostic of knockin. G. GFP expression after EB differentiation of a targeted clone.
Figure 6. Effect of homology arm length for Cas9-mediated knockin.
A. GFP knockin at Oct4 C-terminus using 50 bp or 200 bp homology arms. Knockin efficiency was measured by FACS for GFP expression. B–C. GFP expression in GFP+ FACS clones by microscopy and western blotting. D. PCR genotyping of individual ESC clones. The PCR product (arrowhead) is diagnostic of knockin at Oct4.
Similar articles
- Genome engineering using the CRISPR-Cas9 system.
Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, Zhang F. Ran FA, et al. Nat Protoc. 2013 Nov;8(11):2281-2308. doi: 10.1038/nprot.2013.143. Epub 2013 Oct 24. Nat Protoc. 2013. PMID: 24157548 Free PMC article. - Precision Targeted Mutagenesis via Cas9 Paired Nickases in Rice.
Mikami M, Toki S, Endo M. Mikami M, et al. Plant Cell Physiol. 2016 May;57(5):1058-68. doi: 10.1093/pcp/pcw049. Epub 2016 Mar 2. Plant Cell Physiol. 2016. PMID: 26936792 Free PMC article. - Inhibition of CRISPR/Cas9-Mediated Genome Engineering by a Type I Interferon-Induced Reduction in Guide RNA Expression.
Machitani M, Sakurai F, Wakabayashi K, Nakatani K, Takayama K, Tachibana M, Mizuguchi H. Machitani M, et al. Biol Pharm Bull. 2017;40(3):272-277. doi: 10.1248/bpb.b16-00700. Biol Pharm Bull. 2017. PMID: 28250269 - Application of CRISPR/Cas9 genome editing to the study and treatment of disease.
Pellagatti A, Dolatshad H, Valletta S, Boultwood J. Pellagatti A, et al. Arch Toxicol. 2015 Jul;89(7):1023-34. doi: 10.1007/s00204-015-1504-y. Epub 2015 Apr 1. Arch Toxicol. 2015. PMID: 25827103 Review. - Optimization of genome editing through CRISPR-Cas9 engineering.
Zhang JH, Adikaram P, Pandey M, Genis A, Simonds WF. Zhang JH, et al. Bioengineered. 2016 Apr;7(3):166-74. doi: 10.1080/21655979.2016.1189039. Bioengineered. 2016. PMID: 27340770 Free PMC article. Review.
Cited by
- Rosa26-targeted sheep gene knock-in via CRISPR-Cas9 system.
Wu M, Wei C, Lian Z, Liu R, Zhu C, Wang H, Cao J, Shen Y, Zhao F, Zhang L, Mu Z, Wang Y, Wang X, Du L, Wang C. Wu M, et al. Sci Rep. 2016 Apr 11;6:24360. doi: 10.1038/srep24360. Sci Rep. 2016. PMID: 27063570 Free PMC article. - Pervasive Genotypic Mosaicism in Founder Mice Derived from Genome Editing through Pronuclear Injection.
Oliver D, Yuan S, McSwiggin H, Yan W. Oliver D, et al. PLoS One. 2015 Jun 8;10(6):e0129457. doi: 10.1371/journal.pone.0129457. eCollection 2015. PLoS One. 2015. PMID: 26053263 Free PMC article. - Controlling Ratios of Plasmid-Based Double Cut Donor and CRISPR/Cas9 Components to Enhance Targeted Integration of Transgenes in Chinese Hamster Ovary Cells.
Shin SW, Kim D, Lee JS. Shin SW, et al. Int J Mol Sci. 2021 Feb 27;22(5):2407. doi: 10.3390/ijms22052407. Int J Mol Sci. 2021. PMID: 33673701 Free PMC article. - A Comprehensive Toolbox for Genome Editing in Cultured Drosophila melanogaster Cells.
Kunzelmann S, Böttcher R, Schmidts I, Förstemann K. Kunzelmann S, et al. G3 (Bethesda). 2016 Jun 1;6(6):1777-85. doi: 10.1534/g3.116.028241. G3 (Bethesda). 2016. PMID: 27172193 Free PMC article. - [Research advances on gene therapy for hemophilia A].
Li X, Zhang J, Zhang L, Cheng T, Zhang X. Li X, et al. Zhonghua Xue Ye Xue Za Zhi. 2015 Jul;36(7):620-5. doi: 10.3760/cma.j.issn.0253-2727.2015.07.022. Zhonghua Xue Ye Xue Za Zhi. 2015. PMID: 26304093 Free PMC article. Chinese. No abstract available.
References
Publication types
MeSH terms
Substances
Grants and funding
- U01 HL098188/HL/NHLBI NIH HHS/United States
- U01HL098188/HL/NHLBI NIH HHS/United States
- T32 HL007572/HL/NHLBI NIH HHS/United States
- U01HL098166/HL/NHLBI NIH HHS/United States
- U01 HL098166/HL/NHLBI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases