Longitudinal analysis of microbial interaction between humans and the indoor environment - PubMed (original) (raw)
. 2014 Aug 29;345(6200):1048-52.
doi: 10.1126/science.1254529.
Daniel P Smith 2, Jarrad Hampton-Marcell 1, Sarah M Owens 3, Kim M Handley 1, Nicole M Scott 1, Sean M Gibbons 4, Peter Larsen 5, Benjamin D Shogan 6, Sophie Weiss 7, Jessica L Metcalf 8, Luke K Ursell 9, Yoshiki Vázquez-Baeza 10, Will Van Treuren 8, Nur A Hasan 11, Molly K Gibson 12, Rita Colwell 11, Gautam Dantas 12, Rob Knight 13, Jack A Gilbert 14
Affiliations
- PMID: 25170151
- PMCID: PMC4337996
- DOI: 10.1126/science.1254529
Longitudinal analysis of microbial interaction between humans and the indoor environment
Simon Lax et al. Science. 2014.
Abstract
The bacteria that colonize humans and our built environments have the potential to influence our health. Microbial communities associated with seven families and their homes over 6 weeks were assessed, including three families that moved their home. Microbial communities differed substantially among homes, and the home microbiome was largely sourced from humans. The microbiota in each home were identifiable by family. Network analysis identified humans as the primary bacterial vector, and a Bayesian method significantly matched individuals to their dwellings. Draft genomes of potential human pathogens observed on a kitchen counter could be matched to the hands of occupants. After a house move, the microbial community in the new house rapidly converged on the microbial community of the occupants' former house, suggesting rapid colonization by the family's microbiota.
Copyright © 2014, American Association for the Advancement of Science.
Figures
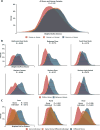
Figure 1. Differentiation in microbial community structure between homes and individuals
Density plots comparing the distributions of weighted UniFrac distances calculated within and between various criteria with accompanying ANOSIM tests of differentiation (all p values are less than 0.0001 based on 10,000 permutations of the randomized dataset). (A) Distribution of distances calculated between two human samples, between two home samples, and between a human sample and a home sample. An ANOSIM test on the effect of source produced a low Rvalue of 0.142, suggesting that home and human surfaces share a large degree of their microbial communities. (B) Distribution of distances for within home and between home comparisons of all samples taken from individual home surfaces. (C) Distribution of distances between human samples for the three sampled surfaces. Comparisons are segregated by whether a sample was compared to another from the same person, to a sample taken from an occupant of the same house, or to a sample taken from a resident of a different home. ANOSIM results are for tests on the effect of the home the sample was taken from (top) and of the individual the sample was taken from (bottom).
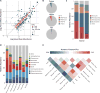
Figure 2. Widespread sharing of microbial taxa between human and home surfaces
(A) Plot of log2-transformed average relative abundances in the human and home environments for all OTUs in the study with greater than 500 reads. OTUs are colored by whether their average relative abundance is significantly different between the home and person environments based on the FDR corrected p-value from a non-parametric t-test run with 1,000 permutations, and are sized based on their log10-transformed number of reads. The dashed line is y=x, indicating an equal average relative abundance. (B) Fraction of all reads from within a source belonging to OTUs shared with other sources, demonstrating the ubiquitous sharing of OTUs between homes and the humans and pets that occupy them. The percent of reads that cluster within source-specific OTUs is less that 0.6% for all three sources. (C) Taxonomic summary of observed relative abundance of abundant phyla across all samples divided by source. (D) Taxonomic summary of observed relative abundance of taxa at class level for all reads in the study by source-specific OTU overlap. (E) Shared phylotypes heatmap for individual surfaces after consolidation of samples taken from the same surface type across temporal sampling series and homes. Pooled samples were rarified to an even depth of 277,500 reads.
Figure 3. Summary of predictive accuracy of Source Tracker models
(A) Percent composition estimate for the correct source for each home surface sample in the study. Samples within each block are ordered by collection date and black boxes occur where a sample is missing because it did not pass quality filtering standards. Across all surfaces, the models averaged a 59% prediction for the correct source. (B) Heatmap of model success across individual surface timeseries. The model was considered to be successful when the proportion of the sink community attributed to the correct source was greater than that attributed to any other source.
Similar articles
- Indoor microbiota in severely moisture damaged homes and the impact of interventions.
Jayaprakash B, Adams RI, Kirjavainen P, Karvonen A, Vepsäläinen A, Valkonen M, Järvi K, Sulyok M, Pekkanen J, Hyvärinen A, Täubel M. Jayaprakash B, et al. Microbiome. 2017 Oct 13;5(1):138. doi: 10.1186/s40168-017-0356-5. Microbiome. 2017. PMID: 29029638 Free PMC article. - Bacterial community assemblages in classroom floor dust of 50 public schools in a large city: characterization using 16S rRNA sequences and associations with environmental factors.
Park JH, Lemons AR, Roseman J, Green BJ, Cox-Ganser JM. Park JH, et al. Microbiome. 2021 Jan 20;9(1):15. doi: 10.1186/s40168-020-00954-2. Microbiome. 2021. PMID: 33472703 Free PMC article. - Microbes and associated soluble and volatile chemicals on periodically wet household surfaces.
Adams RI, Lymperopoulou DS, Misztal PK, De Cassia Pessotti R, Behie SW, Tian Y, Goldstein AH, Lindow SE, Nazaroff WW, Taylor JW, Traxler MF, Bruns TD. Adams RI, et al. Microbiome. 2017 Sep 26;5(1):128. doi: 10.1186/s40168-017-0347-6. Microbiome. 2017. PMID: 28950891 Free PMC article. - The roles of the outdoors and occupants in contributing to a potential pan-microbiome of the built environment: a review.
Leung MH, Lee PK. Leung MH, et al. Microbiome. 2016 May 24;4(1):21. doi: 10.1186/s40168-016-0165-2. Microbiome. 2016. PMID: 27216717 Free PMC article. Review. - House dust microbiome and human health risks.
Shan Y, Wu W, Fan W, Haahtela T, Zhang G. Shan Y, et al. Int Microbiol. 2019 Sep;22(3):297-304. doi: 10.1007/s10123-019-00057-5. Epub 2019 Jan 14. Int Microbiol. 2019. PMID: 30811000 Review.
Cited by
- Limited microbiome differences in captive and semi-wild primate populations consuming similar diets.
Kuthyar S, Watson K, Huang S, Brent LJN, Platt M, Horvath J, Gonzalez-Martinez J, Martínez M, Godoy-Vitorino F, Knight R, Dominguez-Bello MG, Amato KR. Kuthyar S, et al. FEMS Microbiol Ecol. 2022 Oct 3;98(10):fiac098. doi: 10.1093/femsec/fiac098. FEMS Microbiol Ecol. 2022. PMID: 36047944 Free PMC article. - Identifying personal microbiomes using metagenomic codes.
Franzosa EA, Huang K, Meadow JF, Gevers D, Lemon KP, Bohannan BJ, Huttenhower C. Franzosa EA, et al. Proc Natl Acad Sci U S A. 2015 Jun 2;112(22):E2930-8. doi: 10.1073/pnas.1423854112. Epub 2015 May 11. Proc Natl Acad Sci U S A. 2015. PMID: 25964341 Free PMC article. - Natural history of the infant gut microbiome and impact of antibiotic treatment on bacterial strain diversity and stability.
Yassour M, Vatanen T, Siljander H, Hämäläinen AM, Härkönen T, Ryhänen SJ, Franzosa EA, Vlamakis H, Huttenhower C, Gevers D, Lander ES, Knip M; DIABIMMUNE Study Group; Xavier RJ. Yassour M, et al. Sci Transl Med. 2016 Jun 15;8(343):343ra81. doi: 10.1126/scitranslmed.aad0917. Sci Transl Med. 2016. PMID: 27306663 Free PMC article. - Dynamics of the Oral Microbiome During Initial Military Training at Fort Benning, Georgia.
Zudock KK, Player R, Ernlund A, Timm CM, English CE, Ellis MW, Tribble DR, Merrell DS, Bennett JW, Millar EV. Zudock KK, et al. Mil Med. 2024 Jul 3;189(7-8):e1753-e1759. doi: 10.1093/milmed/usad488. Mil Med. 2024. PMID: 38243767 Free PMC article. - Insights into the pan-microbiome: skin microbial communities of Chinese individuals differ from other racial groups.
Leung MH, Wilkins D, Lee PK. Leung MH, et al. Sci Rep. 2015 Jul 16;5:11845. doi: 10.1038/srep11845. Sci Rep. 2015. PMID: 26177982 Free PMC article.
References
- Höppe P, Martinac I. Indoor climate and air quality. Review of current and future topics in the field of ISB study group 10. Int J Biometeorol. 1998;42:1–7. - PubMed
- Custovic A, Taggart SCO, Woodcock A. House duet mite and cat allergen in different indoor environments. Clin Exp Allergy. 1994;24:1164–1168. - PubMed
- Dekio I, et al. Detection of potentially novel bacterial components of the human skin microbiota using culture-independent molecular profiling. J Med Microbiol. 2005;54:1231–8. - PubMed
Publication types
MeSH terms
Grants and funding
- DP2 DK098089/DK/NIDDK NIH HHS/United States
- P01 DK078669/DK/NIDDK NIH HHS/United States
- T32 GM008759/GM/NIGMS NIH HHS/United States
- DP2-DK-098089/DK/NIDDK NIH HHS/United States
- HHMI/Howard Hughes Medical Institute/United States
- U01 HG004866/HG/NHGRI NIH HHS/United States
- R01 HG004872/HG/NHGRI NIH HHS/United States
- T32 GM142607/GM/NIGMS NIH HHS/United States
- T32 EB009412/EB/NIBIB NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical