Phage-host interplay: examples from tailed phages and Gram-negative bacterial pathogens - PubMed (original) (raw)
Review
Phage-host interplay: examples from tailed phages and Gram-negative bacterial pathogens
Soraya Chaturongakul et al. Front Microbiol. 2014.
Abstract
Complex interactions between bacteriophages and their bacterial hosts play significant roles in shaping the structure of environmental microbial communities, not only by genetic transduction but also by modification of bacterial gene expression patterns. Survival of phages solely depends on their ability to infect their bacterial hosts, most importantly during phage entry. Successful dynamic adaptation of bacteriophages when facing selective pressures, such as host adaptation and resistance, dictates their abundance and diversification. Co-evolution of the phage tail fibers and bacterial receptors determine bacterial host ranges, mechanisms of phage entry, and other infection parameters. This review summarizes the current knowledge about the physical interactions between tailed bacteriophages and bacterial pathogens (e.g., Salmonella enterica and Pseudomonas aeruginosa) and the influences of the phage on host gene expression. Understanding these interactions can offer insights into phage-host dynamics and suggest novel strategies for the design of bacterial pathogen biological controls.
Keywords: bacteriophage; host–phage dynamics; host–phage interaction; microbial community; phage resistance mechanism.
Figures
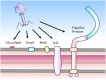
FIGURE 1
Receptors of Salmonella phages. Phages can use a number of cell surface moieties as receptors, including glycolipids (O- and Vi-antigens), integral membrane proteins (e.g., OmpF, BtuB, and TolC) and flagella proteins (FliC, FljB, and FliK).
FIGURE 2
Electron microscopy (EM) of phages. (A) An electron density map of a major capsid protein (MCP) from Bordetella pertussis phage BPP1 determined by single particle reconstruction at 3.5 Angstrom resolution. (B) A ribbon model of BPP1 MCP (Image rendered from deposited structure, EMD-5766 and PDB ID 3J4U; Zhang et al., 2013), (C) a top cartoon shows different stages of T7 phage infection derived from electron cryotomography based on Hu et al. (2013); bottom shows surface rendition of electron density map generated from electron cryotomography and subtomogram averaging (Images rendered from deposited EMDB structures, EMD-5534, EMD-5535, EMD-5536, EMD-5537; Hu et al., 2013).
Similar articles
- Collateral sensitivity increases the efficacy of a rationally designed bacteriophage combination to control Salmonella enterica.
Acton L, Pye HV, Thilliez G, Kolenda R, Matthews M, Turner AK, Yasir M, Holden E, Al-Khanaq H, Webber M, Adriaenssens EM, Kingsley RA. Acton L, et al. J Virol. 2024 Mar 19;98(3):e0147623. doi: 10.1128/jvi.01476-23. Epub 2024 Feb 20. J Virol. 2024. PMID: 38376991 Free PMC article. - Molecular Basis of Bacterial Host Interactions by Gram-Positive Targeting Bacteriophages.
Dunne M, Hupfeld M, Klumpp J, Loessner MJ. Dunne M, et al. Viruses. 2018 Jul 28;10(8):397. doi: 10.3390/v10080397. Viruses. 2018. PMID: 30060549 Free PMC article. Review. - A Novel Group of Promiscuous Podophages Infecting Diverse Gammaproteobacteria from River Communities Exhibits Dynamic Intergenus Host Adaptation.
Cazares D, Cazares A, Figueroa W, Guarneros G, Edwards RA, Vinuesa P. Cazares D, et al. mSystems. 2021 Feb 2;6(1):e00773-20. doi: 10.1128/mSystems.00773-20. mSystems. 2021. PMID: 33531404 Free PMC article. - Understanding bacteriophage specificity in natural microbial communities.
Koskella B, Meaden S. Koskella B, et al. Viruses. 2013 Mar 11;5(3):806-23. doi: 10.3390/v5030806. Viruses. 2013. PMID: 23478639 Free PMC article. Review. - Bacteriophage specificity is impacted by interactions between bacteria.
Bisesi AT, Möbius W, Nadell CD, Hansen EG, Bowden SD, Harcombe WR. Bisesi AT, et al. mSystems. 2024 Mar 19;9(3):e0117723. doi: 10.1128/msystems.01177-23. Epub 2024 Feb 20. mSystems. 2024. PMID: 38376179 Free PMC article.
Cited by
- Comparative Genomics of Six Lytic Bacillus subtilis Phages from the Southwest United States.
Vill AC, Delesalle VA, Tomko BE, Lichty KB, Strine MS, Guffey AA, Burton EA, Tanke NT, Krukonis GP. Vill AC, et al. Phage (New Rochelle). 2022 Sep 1;3(3):171-178. doi: 10.1089/phage.2022.0030. Epub 2022 Sep 19. Phage (New Rochelle). 2022. PMID: 36793550 Free PMC article. - Nine Novel Phages from a Plateau Lake in Southwest China: Insights into Aeromonas Phage Diversity.
Bai M, Cheng YH, Sun XQ, Wang ZY, Wang YX, Cui XL, Xiao W. Bai M, et al. Viruses. 2019 Jul 5;11(7):615. doi: 10.3390/v11070615. Viruses. 2019. PMID: 31284428 Free PMC article. - Metagenomic analysis of wastewater phageome from a University Hospital in Turkey.
Salih H, Karaynir A, Yalcin M, Oryasin E, Holyavkin C, Basbulbul G, Bozdogan B. Salih H, et al. Arch Microbiol. 2022 May 30;204(6):353. doi: 10.1007/s00203-022-02962-2. Arch Microbiol. 2022. PMID: 35637399 - Bacteriophage Cocktail Can Effectively Control Salmonella Biofilm in Poultry Housing.
Korzeniowski P, Śliwka P, Kuczkowski M, Mišić D, Milcarz A, Kuźmińska-Bajor M. Korzeniowski P, et al. Front Microbiol. 2022 Jun 29;13:901770. doi: 10.3389/fmicb.2022.901770. eCollection 2022. Front Microbiol. 2022. PMID: 35847069 Free PMC article. - Low-level predation by lytic phage phiIPLA-RODI promotes biofilm formation and triggers the stringent response in Staphylococcus aureus.
Fernández L, González S, Campelo AB, Martínez B, Rodríguez A, García P. Fernández L, et al. Sci Rep. 2017 Jan 19;7:40965. doi: 10.1038/srep40965. Sci Rep. 2017. PMID: 28102347 Free PMC article.
References
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources