Crystal structure of the γ-secretase component nicastrin - PubMed (original) (raw)
Crystal structure of the γ-secretase component nicastrin
Tian Xie et al. Proc Natl Acad Sci U S A. 2014.
Abstract
γ-Secretase is an intramembrane protease responsible for the generation of amyloid-β (Aβ) peptides. Aberrant accumulation of Aβ leads to the formation of amyloid plaques in the brain of patients with Alzheimer's disease. Nicastrin is the putative substrate-recruiting component of the γ-secretase complex. No atomic-resolution structure had been identified on γ-secretase or any of its four components, hindering mechanistic understanding of γ-secretase function. Here we report the crystal structure of nicastrin from Dictyostelium purpureum at 1.95-Å resolution. The extracellular domain of nicastrin contains a large lobe and a small lobe. The large lobe of nicastrin, thought to be responsible for substrate recognition, associates with the small lobe through a hydrophobic pivot at the center. The putative substrate-binding pocket is shielded from the small lobe by a lid, which blocks substrate entry. These structural features suggest a working model of nicastrin function. Analysis of nicastrin structure provides insights into the assembly and architecture of the γ-secretase complex.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Fig. 1.
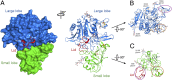
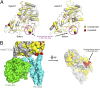
Structure of the nicastrin ECD from D. Purpureum. (A) The overall structure of the nicastrin ECD from D. Purpureum is shown in surface representation (Left) and ribbon diagram (Right). The structure can be divided into a large lobe (blue) and a small lobe (green). An extended loop from the small lobe (red) forms a lid to cover an otherwise exposed surface region on the large lobe. The highly conserved Trp145 in the lid is indicated in red ball-and-stick representation. Five N-linked glycans are displayed in light gray; six disulfide bonds are shown in orange. All structural figures were prepared with PyMOL (42). (B) Structure of the large lobe of nicastrin. The large lobe contains a core and a number of additional structural motifs on the surface, notably a pair of antiparallel β-strands (purple oval) and a small globular domain (orange oval). (C) The structure of the small lobe of nicastrin. As does the large lobe, the small lobe contains two prominent structural elements beyond the core: a small globular domain (orange oval) and a lid that interacts with the large lobe (purple oval).
Fig. 2.
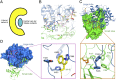
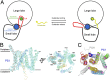
A unique pattern of interactions at the interface between the large and small lobes of DpNCT. (A) A schematic diagram of the interface between the large and small lobes of nicastrin. At the center of the interface is a high density of van der Waals interactions. At the periphery of the interface are a number of H-bonds that appear in a C-shaped distribution. These interactions are conserved between HsNCT and DpNCT. (B) A close-up view of the H-bonds between the large and small lobes of nicastrin. The lid from the small lobe is highlighted in red. H-bonds are represented by red dashed lines. The same coloring scheme is used in all figures. (C) A close-up view of the hydrophobic pivot at the center of the interface between the large and small lobes of nicastrin. Phe244 and Phe245 from the large lobe are nestled in a greasy pocket formed by hydrophobic residues from the small lobe. (D) A close-up view of the interactions between Trp145 from the lid of the small lobe and residues from the large lobe.
Fig. 3.
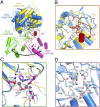
Identification and features of the putative substrate-binding pocket in DpNCT. (A) Structural comparison between BAP (PDB ID code 2EK9) and nicastrin. Overlaying the large lobe of BAP with that of nicastrin reveals quite different positions for their small lobes, which are separated from each other by a rotation of ∼100°. The large and small lobes of BAP are colored yellow and magenta, respectively. (B) Identification of the putative substrate-binding pocket in nicastrin. The structure of BAP is superimposed here to illustrate its substrate-binding pocket, which is occupied by the competitive inhibitor BES (orange). The two bound zinc ions in BAP are shown as gray spheres. The putative substrate-binding pocket in nicastrin is located in the large lobe and is shielded by the lid from the small lobe. (C) A close-up view of zinc coordination in BAP. The nicastrin structure is superimposed here for comparison. The two zinc ions in BAP are missing in nicastrin. The residues that coordinate the zinc ions in BAP are colored magenta; the corresponding residues in nicastrin are shown in gray. (D) A close-up view of the putative substrate-binding pocket in the large lobe of nicastrin. These residues are mostly polar. The amino acids Glu289 (corresponding to Glu333 in HsNCT) and Tyr293 (corresponding to Tyr337 of the DYIGS motif in HsNCT) are both located in the putative substrate-binding pocket.
Fig. 4.
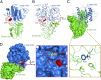
Structural features of the modeled HsNCT. (A) Overall structure of the modeled HsNCT. The atomic model for HsNCT was generated on the basis of the crystal structure of DpNCT and fitted into the 4.5-Å resolution EM density of human γ-secretase (26). The large and small lobes of HsNCT model are colored blue and green, respectively. The lid from the small lobe is highlighted in red. (B) The modeled structure of HsNCT contains important previously unidentified features compared with the previous partial model. These features include two small globular domains, one in each lobe, and a lid from the small lobe that covers the putative substrate-binding site on the large lobe. The previous partial model of HsNCT is colored gray. (C) A close-up view of the hydrophobic pivot at the center of the interface between the large and small lobes of HsNCT. Phe287 from the large lobe interacts with four hydrophobic residues from the small lobe. These residues are highly conserved in different organisms. (D) A close-up view of the lid from the small domain of HsNCT. As is Trp145 in DpNCT, Trp164 in HsNCT is located in the lid of the small lobe, making contacts with residues in the large lobe.
Fig. 5.
Structural conservation between DpNCT and HsNCT. (A) Comparison of conserved structural features in DpNCT and HsNCT. Invariant residues among all five organisms (Fig. S2) are highlighted in red; conserved amino acids are colored yellow. (B) The bottom surface of the small lobe, which is in contact with the TMs of γ-secretase, is highly conserved. The small globular domain from the small lobe, which is in contact with the thin end of the TMs, is particularly conserved. The TMs of γ-secretase are shown in surface representation.
Fig. 6.
Implications for nicastrin function and reassignment of the TMs in PS1. (A) A working model of nicastrin. In this hypothetical model, binding the lid to the large lobe blocks access to the substrate-binding site. Rotation of the large lobe relative to the small lobe around the central hydrophobic pivot may cause the lid to open, exposing the putative substrate-binding site and allowing substrate access. (B) Reassignment of the TMs in PS1 of human γ-secretase. The EM density for TM2 and TM6 of PS1 was relatively poor. The EM density for the seven remaining TMs of PS1 was clearly assigned. (C) Structural comparison of PS1 (rainbow) with the archaeal homolog mmPSH (gray). The clearly assigned seven TMs of PS1 (TM1, TM3, TM4, TM5, TM7, TM8, and TM9) superimpose well with the corresponding TMs of mmPSH.
Comment in
- Structure of nicastrin unveils secrets of γ-secretase.
Bolduc DM, Wolfe MS. Bolduc DM, et al. Proc Natl Acad Sci U S A. 2014 Oct 14;111(41):14643-4. doi: 10.1073/pnas.1416637111. Epub 2014 Sep 29. Proc Natl Acad Sci U S A. 2014. PMID: 25267656 Free PMC article. No abstract available.
Similar articles
- Identification of a tetratricopeptide repeat-like domain in the nicastrin subunit of γ-secretase using synthetic antibodies.
Zhang X, Hoey RJ, Lin G, Koide A, Leung B, Ahn K, Dolios G, Paduch M, Ikeuchi T, Wang R, Li YM, Koide S, Sisodia SS. Zhang X, et al. Proc Natl Acad Sci U S A. 2012 May 29;109(22):8534-9. doi: 10.1073/pnas.1202691109. Epub 2012 May 14. Proc Natl Acad Sci U S A. 2012. PMID: 22586122 Free PMC article. - Structure of the transmembrane domain of human nicastrin-a component of γ-secretase.
Li Y, Liew LS, Li Q, Kang C. Li Y, et al. Sci Rep. 2016 Jan 18;6:19522. doi: 10.1038/srep19522. Sci Rep. 2016. PMID: 26776682 Free PMC article. - Glu(332) in the Nicastrin ectodomain is essential for gamma-secretase complex maturation but not for its activity.
Chávez-Gutiérrez L, Tolia A, Maes E, Li T, Wong PC, de Strooper B. Chávez-Gutiérrez L, et al. J Biol Chem. 2008 Jul 18;283(29):20096-105. doi: 10.1074/jbc.M803040200. Epub 2008 May 23. J Biol Chem. 2008. PMID: 18502756 - Nicastrin: gatekeeper of the gamma-secretase complex.
De Strooper B. De Strooper B. Cell. 2005 Aug 12;122(3):318-20. doi: 10.1016/j.cell.2005.07.021. Cell. 2005. PMID: 16096051 Review. - Gamma-secretase: structure, function, and modulation for Alzheimer's disease.
Wolfe MS. Wolfe MS. Curr Top Med Chem. 2008;8(1):2-8. doi: 10.2174/156802608783334024. Curr Top Med Chem. 2008. PMID: 18220927 Review.
Cited by
- Identification of domains in Plasmodium falciparum proteins of unknown function using DALI search on AlphaFold predictions.
Behrens HM, Spielmann T. Behrens HM, et al. Sci Rep. 2024 May 8;14(1):10527. doi: 10.1038/s41598-024-60058-x. Sci Rep. 2024. PMID: 38719885 Free PMC article. - Analysis of nicastrin gene phylogeny and expression in zebrafish.
Lim A, Moussavi Nik SH, Ebrahimie E, Lardelli M. Lim A, et al. Dev Genes Evol. 2015 Jun;225(3):171-8. doi: 10.1007/s00427-015-0500-9. Epub 2015 May 5. Dev Genes Evol. 2015. PMID: 25940938 - γ-secretase promotes Drosophila postsynaptic development through the cleavage of a Wnt receptor.
Restrepo LJ, DePew AT, Moese ER, Tymanskyj SR, Parisi MJ, Aimino MA, Duhart JC, Fei H, Mosca TJ. Restrepo LJ, et al. Dev Cell. 2022 Jul 11;57(13):1643-1660.e7. doi: 10.1016/j.devcel.2022.05.006. Epub 2022 Jun 1. Dev Cell. 2022. PMID: 35654038 Free PMC article. - Structure and Function of the γ-Secretase Complex.
Wolfe MS. Wolfe MS. Biochemistry. 2019 Jul 9;58(27):2953-2966. doi: 10.1021/acs.biochem.9b00401. Epub 2019 Jun 25. Biochemistry. 2019. PMID: 31198028 Free PMC article. - Non-small-cell lung carcinoma: role of the Notch signaling pathway.
Barse L, Bocchetta M. Barse L, et al. Lung Cancer (Auckl). 2015 Jun 26;6:43-53. doi: 10.2147/LCTT.S60329. eCollection 2015. Lung Cancer (Auckl). 2015. PMID: 28210150 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources