PKD1 phosphorylation-dependent degradation of SNAIL by SCF-FBXO11 regulates epithelial-mesenchymal transition and metastasis - PubMed (original) (raw)
PKD1 phosphorylation-dependent degradation of SNAIL by SCF-FBXO11 regulates epithelial-mesenchymal transition and metastasis
Hanqiu Zheng et al. Cancer Cell. 2014.
Abstract
Metastatic dissemination is often initiated by the reactivation of an embryonic development program referred to as epithelial-mesenchymal transition (EMT). The transcription factor SNAIL promotes EMT and elicits associated pathological characteristics such as invasion, metastasis, and stemness. To better understand the posttranslational regulation of SNAIL, we performed a luciferase-based, genome-wide E3 ligase siRNA library screen and identified SCF-FBXO11 as an important E3 that targets SNAIL for ubiquitylation and degradation. Furthermore, we discovered that SNAIL degradation by FBXO11 is dependent on Ser-11 phosphorylation of SNAIL by protein kinase D1 (PKD1). FBXO11 blocks SNAIL-induced EMT, tumor initiation, and metastasis in multiple breast cancer models. These findings establish the PKD1-FBXO11-SNAIL axis as a mechanism of posttranslational regulation of EMT and cancer metastasis.
Copyright © 2014 Elsevier Inc. All rights reserved.
Figures
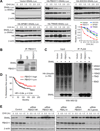
Figure 1. A genome-wide functional screen for E3 ubiquitin ligase(s) targeting SNAIL protein for degradation
(A) In SUM1315 cells, endogenous SNAIL protein was detected by western blot after CHX (left panel) or MG132 treatment (right panel) for the indicated hours. Western blot data was quantified in Figure S1A. (B) Illustration of dual-luciferase reporter screening system for SNAIL-targeting E3 ligases. (C, D) CHX pulse-chase experiment demonstrated the degradation of SNAIL-F-Luc fusion protein in SUM-SNAIL-Luc/R-Luc cells. Western blot data was quantified in (D). Data is presented as mean ± SEM. (E) Degradation of SNAIL-F-Luc protein was monitored by normalized luciferase activity measurement. Data is presented as mean ± SEM. (F) Experimental procedure flow chart for identification of the E3 ubiquitin ligase(s) targeting SNAIL protein. See text for details. (G) Luciferase based siRNA library screen against human E3 ligases identified multiple E3 candidates, when knocked down in SUM-SNAIL-Luc/R-Luc cells, increased luciferase activity by more than 2 fold (upper panel). A second round of siRNA screening (lower left panel) and immunoblotting (lower right panel) was performed for confirmation of candidates. (H, I) 293T cells were transfected according to each panel labeling. Co-IP experiment was performed using either an HA antibody to pull down HA-tagged E3 ligase proteins (H) or an anti-FLAG antibody against SNAIL-FLAG protein. See also Figure S1 and Table S1 and S2.
Figure 2. FBXO11 is a bona fide E3 ligase targeting SNAIL protein for ubiquitination and degradation
(A) Four E3 ligase candidates or control pLEX-vector were co-transfected with SNAIL-Luc plasmid into 293T cells and CHX pulse-chase assay was performed. Western blot data was quantified using ImageJ software. (B) LM2 cells were treated with 10 µM MG132 for 6 hr before the cell lysate was immunoprecipitated with FBXO11 antibody or IgG control and subjected to western blot analysis. (C) 293T cells were co-transfected by HA-Ub, SNAIL-Flag, together with vector, FBXO11 or FBXO11 - ΔF. Cells were treated with MG132 for 6 hr before cell lysates were immunoprecipitated using a denature IP protocol to pull down SNAIL protein, and the polyubiquitylated SNAIL protein was detected by anti-HA antibody. (D) Kaplan-Meier plots of metastasis-free survival of patients stratified by expression of FBXO11. Data obtained from the NKI breast cancer dataset (van de Vijver et al., 2002). (E) MCF10A cells were transfected with indicated siRNAs, followed by the CHX pulse-chase assay. Quantification data is shown in Figure S2P See also Figure S2.
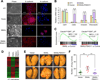
Figure 3. FBXO11 blocks SNAIL-induced EMT and reduces tumorigenesis capability in HMLEN cells
(A) Phase contrast and immunofluorescent images of HMLEN cells with stable overexpression of SNAIL or SNAIL together with FBXO11. Scale bar, 100 µm in phase contrast images and 25 µm in IF images. (B) GSEA analysis of enriconment of EMT gene signature in HMLEN cells after overexpression of SNAIL with or without FBXO11. (C) Heatmap was generated using hierarchical clustering of the microarray data for the indicated HMLEN cell lines. The 2327 gene probes used for clustering were those showing >2 folds expression changes upon SNAIL overexpression. Experiment was performed in duplicates. (D) Boyden chamber invasion assay of HMLEN cells with SNAIL or SNAIL/FBXO11 overexpression. The data were shown as mean of collected data from 3 triplicate wells of three independent experiments. Data is presented as mean ± SEM, **p < 0.01, ***p < 0.001. (E) Quantification of colony formation assay results for the indicated HMLEN cell lines. Data is presented as mean ± SEM, **p < 0.01, ***p < 0.001. (F, G) 2×104 of the indicated HMLEN cells were orthotopically injected into nude mice (n = 8) and primary mammary tumor growth was measured weekly after injection, with representative tumor images shown in (G). Data represent mean ± s.d **p < 0.01 by two-tailed Student’s t test. See also Figure S3.
Figure 4. FBXO11 blocked SNAIL-induced EMT and metastasis in 4T1 breast cancer cell line
(A) Phase contrast and IF images for 4T1 cells transduced with indicated lentiviruses. SNAIL-induced strong EMT program in 4T1 cells with downregulation of E-cadherin and upregulation of N-cadherin expression, while FBXO11 blocked SNAIL-induced EMT. Scale bars, 25µm. (B) qRT-PCR analysis of EMT marker mRNAs in the indicated 4T1 cell lines. Results were normalized to GAPDH mRNA level. Data represent average ± SEM, *p < 0.05 by Student's t test. (**C**) GSEA analysis demonstrated FBXO11 expression inhibited SNAIL induced EMT gene signature in 4T1 cells. (**D**) Heatmap was generated using hierarchical clustering. The 1400 gene probes used for clustering were those showing >2 folds expression changes in SNAIL overexpressing cells. (E) 105 cells was orthotopically injected into Balb/c mice (n = 10). Mice were sacrificed 3 weeks later. Representative lung metastasis nodule images were presented. (F) Numbers of lung metastasis lesions of mice injected with the indicated 4T1 cell lines. **p < 0.01 by Mann–Whitney U test. See also Figure S4.
Figure 5. Knockdown of endogenous FBXO11 promotes breast cancer metastasis
(A) Real-time PCR analysis of Fbxo11, Cdh1 and Vimentin expression in the parental and control 4T1 cell line, and two Fbxo11-KD cell lines. Data represent mean ± SD. **: p < 0.01 by two-tailed Student’s t test. (B) Western blot analysis of Snail, E-cadherin and Vimentin expression in the indicated 4T1 cell lines. (C) Phase contrast images and immunofluorescent images of EMT markers in the indicated 4T1 cell lines. Scale bars, 25µm. (D) 105 tumor cells from various 4T1 cell lines were orthotopically injected into Balb/c mice and primary tumors were removed 10 days later. Lung metastasis nodules were counted after sacrificing the mice at 38 days post injection. **p < 0.01 by Mann–Whitney U test. (E) Representative lung metastasis nodule images were presented. See also Figure S5.
Figure 6. Serine-11 in the SNAG domain of SNAIL is responsible for interaction with FBXO11
(A) 293T cells were transfected with SNAIL-Flag and HA-FBXO11 for 2 days. Cell lysates were collected and treated either with or without CIP for 1 hour before subjected to co-IP experiment. (B) Schematic representation of SNAIL protein structure and the deletion mutation constructs generated to map out the interaction domain for SNAIL-FBXO11 interaction. (C) Wild-type FLAG-tagged SNAIL and SNAIL truncation mutants were co-transfected with HA-FBXO11 into 293T cells, cell lysates were collected for co-IP experiment using anti-FLAG antibody. (D) Wild-type SNAIL and SNAIL–S11E, SNAIL-S11V single amino acid mutants were co-transfected with HA-FBXO11 into 293T cells, cell lysates were collected for co-IP experiment using anti-HA antibody. (E) CHX pulse-chase assay for wild-type SNAIL, SNAIL-S11E, and SNAIL-S11V in the absence of exogenously expressed FBXO11 in 293T cells. Data was quantified using ImageJ software. (F) Western blot images of experiments in (E). (G) 293T cells were transfected with the indicated expression plasmids and cell lysates were collected for immunoblotting two days after transfection. (H, I) 293T stable cells expressing corresponding wild type SNAIL or mutant SNAIL were transfected with the indicated FBXO11 expression plasmids. Pulse-chase assay was performed 2 days later. Data was quantified in (I) using ImageJ software. See also Figure S6.
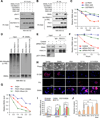
Figure 7. PKD1 dependent phosphorylation of SNAIL Ser-11 is critical for SNAIL protein degradation
(A) Detection of Ser11 phosphorylation of immunoprecipitated SNAIL protein when SNAIL was co-transfected with indicated plasmids or treated with PKD1 inhibitor CID755673 in 293T cells. (B) 293T cells were transfected or treated similarly to (A) and then lysed, and co-IP experiment was performed using anti-HA antibody. (C) In 293T cells stably transfected with SNAIL, SNAIL protein turnover rates were determined by CHX pulse-chase assays after cells were transfected with FBXO11 together with the indicated plasmids or inhibitor CID755673. Western blot images in Figure S8A were quantified using Image J software. (D) 293T cells were transfected with the indicated plasmids. Cells were treated with DMSO or PKD1 kinase inhibitor CID755673 before lysing in denature lysis buffer. Cell lyslates were subjected to denature IP with SNAIL antibody. Poly-ubiquitylated SNAIL protein was visualized by HA antibody blotting against HA-Ubiquitin. (E) MCF10A cells were treated with either 10nM control or PKD1 -targeting siRNA before subjected to co-IP and western blot analysis using the indicated antibodies. In (A), (B), (D) and (E), cells were treated with MG132 for 6 hr before co-IP experiments. (F) MCF10A cells were transfected with control or two PKD1 siRNAs. Two days later, endogenous SNAIL degradation was determined by CHX pulse-chase assay. Western blot images in Figure S7E were quantified using Image J software. (G) 293T-SNAIL stable cell line was transfected with HA-PKD1, together with either vector or GST-tagged constitutively active RhoA-CA, and CHX pulse-chase assay was performed for SNAIL. Western blot images in Figure S7H were quantified using Image J software. (H) Representative phase contrast images and immunofluorescent images of MCF7 cells stably expressing SNAIL or SNAIL mutants. Scale bars: 25µm. (I) qRT-PCR was performed for CDH1, CDH2, VIM, and FN in the indicated MCF7 stable cell lines. Data represent mean ± s.d., *p < 0.05 by two-tailed Student’s t test. (J) Boyden chamber invasion assay of MCF7 cells stably expressing SNAIL or SNAIL stabilization mutants. **p < 0.01 by Student's t test. See also Figure S7.
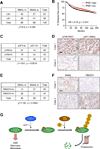
Figure 8. SNAIL protein expression level is positively correlated with lymph node invasion and negatively correlated with FBXO11 expression level in breast cancer patients
(A) Correlation study of SNAIL expression level with lymph node invasion in 136 breast tumor specimens. SNAIL-lo: SNAIL IHC staining lower than median; SNAIL-hi: SNAIL IHC staining higher than median; LN-: without lymph node invasion; LN+: with lymph node invasion. χ2=4.9, p = 0.026 by chi-square test. (B) Kaplan-Meier plots of distant relapse-free survival of patients, stratified by expression of PKD1. Data obtained from the Kaplan-Meier plotter database (Gyorffy et al., 2010). (C) Correlation study of activated PKD1 (pY95-PKD1) and S11-SNAIL in a breast tumor tissue microarray (US Biomax BC081120), χ2=10.0, p = 0.0016 by chi-square test. (D) Representative IHC images of pY95-PKD1 and pS11 -SNAIL in breast tumors. Scale bars, 100µm. (E) Correlation study of SNAIL and FBXO11 in the same breast tumor tissue microarray. χ2=7.49, p = 0.0062 by chi-square test. (F) Representative IHC images of SNAIL and FBXO11 in breast tumors. Scale bars: 100µm. (G) Schematic model for PKD1-dependent SNAIL protein ubiquitylation and degradation by FBXO11. SNAIL protein is first phosphorylated by PKD1 kinase at Serine-11 residue before it can be recognized and ubiquitylated by SCF-FBXO11 E3 ligase complex. Poly-ubiquitylated SNAIL protein is then degraded through 26S proteosomal degradation pathway.
Similar articles
- FBXO11 promotes ubiquitination of the Snail family of transcription factors in cancer progression and epidermal development.
Jin Y, Shenoy AK, Doernberg S, Chen H, Luo H, Shen H, Lin T, Tarrash M, Cai Q, Hu X, Fiske R, Chen T, Wu L, Mohammed KA, Rottiers V, Lee SS, Lu J. Jin Y, et al. Cancer Lett. 2015 Jun 28;362(1):70-82. doi: 10.1016/j.canlet.2015.03.037. Epub 2015 Mar 28. Cancer Lett. 2015. PMID: 25827072 Free PMC article. - FBXO11 targets BCL6 for degradation and is inactivated in diffuse large B-cell lymphomas.
Duan S, Cermak L, Pagan JK, Rossi M, Martinengo C, di Celle PF, Chapuy B, Shipp M, Chiarle R, Pagano M. Duan S, et al. Nature. 2012 Jan 5;481(7379):90-3. doi: 10.1038/nature10688. Nature. 2012. PMID: 22113614 Free PMC article. - F-box proteins: Keeping the epithelial-to-mesenchymal transition (EMT) in check.
Díaz VM, de Herreros AG. Díaz VM, et al. Semin Cancer Biol. 2016 Feb;36:71-9. doi: 10.1016/j.semcancer.2015.10.003. Epub 2015 Nov 11. Semin Cancer Biol. 2016. PMID: 26506454 Review. - FBXO22, an epigenetic multiplayer coordinating senescence, hormone signaling, and metastasis.
Johmura Y, Harris AS, Ohta T, Nakanishi M. Johmura Y, et al. Cancer Sci. 2020 Aug;111(8):2718-2725. doi: 10.1111/cas.14534. Epub 2020 Jul 17. Cancer Sci. 2020. PMID: 32536008 Free PMC article. Review.
Cited by
- Integrative Omics Uncovers Low Tumorous Magnesium Content as A Driver Factor of Colorectal Cancer.
Zhang R, Hu M, Liu Y, Li W, Xu Z, He S, Lu Y, Gong Y, Wang X, Hai S, Li S, Qi S, Li Y, Shu Y, Du D, Zhang H, Xu H, Zhou Z, Lei P, Chen HN, Dai L. Zhang R, et al. Genomics Proteomics Bioinformatics. 2024 Oct 15;22(4):qzae053. doi: 10.1093/gpbjnl/qzae053. Genomics Proteomics Bioinformatics. 2024. PMID: 39052867 Free PMC article. - The Post-Translational Regulation of Epithelial-Mesenchymal Transition-Inducing Transcription Factors in Cancer Metastasis.
Kang E, Seo J, Yoon H, Cho S. Kang E, et al. Int J Mol Sci. 2021 Mar 30;22(7):3591. doi: 10.3390/ijms22073591. Int J Mol Sci. 2021. PMID: 33808323 Free PMC article. Review. - Secreted TGF-beta-induced protein promotes aggressive progression in bladder cancer cells.
Zou J, Huang R, Li H, Wang B, Chen Y, Chen S, Ou K, Wang X. Zou J, et al. Cancer Manag Res. 2019 Jul 25;11:6995-7006. doi: 10.2147/CMAR.S208984. eCollection 2019. Cancer Manag Res. 2019. PMID: 31440088 Free PMC article. - Ubiquitin-specific peptidase 37: an important cog in the oncogenic machinery of cancerous cells.
Chauhan R, Bhat AA, Masoodi T, Bagga P, Reddy R, Gupta A, Sheikh ZA, Macha MA, Haris M, Singh M. Chauhan R, et al. J Exp Clin Cancer Res. 2021 Nov 10;40(1):356. doi: 10.1186/s13046-021-02163-7. J Exp Clin Cancer Res. 2021. PMID: 34758854 Free PMC article. Review. - STK39 promotes breast cancer invasion and metastasis by increasing SNAI1 activity upon phosphorylation.
Qiu Z, Dong B, Guo W, Piotr R, Longmore G, Yang X, Yu Z, Deng J, Evers BM, Wu Y. Qiu Z, et al. Theranostics. 2021 Jun 11;11(16):7658-7670. doi: 10.7150/thno.62406. eCollection 2021. Theranostics. 2021. PMID: 34335956 Free PMC article.
References
- Batlle E, Sancho E, Franci C, Dominguez D, Monfar M, Baulida J, Garcia De Herreros A. The transcription factor snail is a repressor of E-cadherin gene expression in epithelial tumour cells. Nat Cell Biol. 2000;2:84–89. - PubMed
- Brabletz T. To differentiate or not--routes towards metastasis. Nature reviews Cancer. 2012;12:425–436. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01CA134519/CA/NCI NIH HHS/United States
- P30 CA072720/CA/NCI NIH HHS/United States
- GM086435/GM/NIGMS NIH HHS/United States
- R01 CA140182/CA/NCI NIH HHS/United States
- R01 CA141062/CA/NCI NIH HHS/United States
- T32 GM007388/GM/NIGMS NIH HHS/United States
- CA140182/CA/NCI NIH HHS/United States
- R01 CA134519/CA/NCI NIH HHS/United States
- R01 GM086435/GM/NIGMS NIH HHS/United States
- R01CA141062/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials