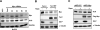The mTORC1/S6K1 pathway regulates glutamine metabolism through the eIF4B-dependent control of c-Myc translation - PubMed (original) (raw)
The mTORC1/S6K1 pathway regulates glutamine metabolism through the eIF4B-dependent control of c-Myc translation
Alfredo Csibi et al. Curr Biol. 2014.
Retraction in
- Retraction Notice to: The mTORC1/S6K1 Pathway Regulates Glutamine Metabolism through the eIF4B- Dependent Control of c-Myc Translation.
Csibi A, Lee G, Yoon SO, Tong H, Ilter D, Elia I, Fendt SM, Roberts TM, Blenis J. Csibi A, et al. Curr Biol. 2025 Jan 6;35(1):232. doi: 10.1016/j.cub.2024.12.016. Epub 2024 Dec 14. Curr Biol. 2025. PMID: 39675354 No abstract available.
Abstract
Growth-promoting signaling molecules, including the mammalian target of rapamycin complex 1 (mTORC1), drive the metabolic reprogramming of cancer cells required to support their biosynthetic needs for rapid growth and proliferation. Glutamine is catabolyzed to α-ketoglutarate (αKG), a tricarboxylic acid (TCA) cycle intermediate, through two deamination reactions, the first requiring glutaminase (GLS) to generate glutamate and the second occurring via glutamate dehydrogenase (GDH) or transaminases. Activation of the mTORC1 pathway has been shown previously to promote the anaplerotic entry of glutamine to the TCA cycle via GDH. Moreover, mTORC1 activation also stimulates the uptake of glutamine, but the mechanism is unknown. It is generally thought that rates of glutamine utilization are limited by mitochondrial uptake via GLS, suggesting that, in addition to GDH, mTORC1 could regulate GLS. Here we demonstrate that mTORC1 positively regulates GLS and glutamine flux through this enzyme. We show that mTORC1 controls GLS levels through the S6K1-dependent regulation of c-Myc (Myc). Molecularly, S6K1 enhances Myc translation efficiency by modulating the phosphorylation of eukaryotic initiation factor eIF4B, which is critical to unwind its structured 5' untranslated region (5'UTR). Finally, our data show that the pharmacological inhibition of GLS is a promising target in pancreatic cancers expressing low levels of PTEN.
Copyright © 2014 Elsevier Ltd. All rights reserved.
Figures
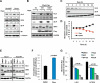
Figure 1. The mTORC1 pathway regulates GLS1
(A-C, E) GLS protein levels in whole cell lysates from: (A) Tsc2 WT and _Tsc2_−/− MEFs treated with rapamycin for 8h; (B) HEK293T cells stably expressing Rheb WT, the mutant S16H Rheb or empty vector (EV) and treated with rapamycin for 24h; (C) _Tsc2_−/− MEFs treated with rapamycin at indicated time points; (E) Tsc2 WT and _Tsc2_−/− MEFs treated with the indicated compounds for 8h. The concentrations of the compounds were: rapamycin 20ng/mL; LY294002 20μM, and BEZ235 10μM. (D) Time course of glutamine consumption in Tsc2−/− MEFs incubated with or without 20ng/mL rapamycin for 24h. Each time data point is an average of triplicate experiments. (F) Intracellular glutamine levels in _Tsc2_−/− MEFs treated with rapamycin for 24h. (G) Glutamine flux in _Tsc2_−/− MEFs expressing an empty vector (EV) or re-expressing TSC2 treated with the indicated compounds for 24h. The concentrations of the compounds were: rapamycin 20ng/mL; LY294002 20μM, and BEZ235 10μM, BPTES 10μM, and DON 1mM. The mean is shown; error bars represent SEM from at least three biological replicates. Numbers below the immunoblot image represent quantification normalized to the loading control. See also Figure S1.
Figure 2. The mTORC1 pathway regulates GLS1 via Myc
(A-C) GLS and Myc protein levels in whole cell lysates from: (A) BxPC3 cells transfected with a non-targeting control siRNA (NTC) or four independent siRNAs against Myc for 72h; (B) Tsc2 WT and _Tsc2_−/− MEFs treated with rapamycin 20ng/mL for 8h; (C) _Tsc2_−/− MEFs stably expressing Myc or empty vector (EV), and treated with rapamycin 20ng/mL for 24h.
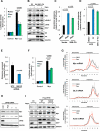
Figure 3. The mTORC1 substrate S6K1 controls GLS through Myc mRNA translation
(A) Normalized luciferase light units of _Tsc2_−/− MEFs stably expressing a Myc-responsive firefly luciferase construct (Myc-Luc) or vector control (pCignal Lenti-TRE Reporter). Myc transcriptional activity was measured after treatment with rapamycin 20ng/mL or PF4708671 10μM for 8h. (B) GLS and Myc protein levels in whole cell lysates from HEK293T cells expressing HA-S6K1-CA (F5A-R3A-T389E) or empty vector (EV) treated with rapamycin 20ng/mL for 24h. (C-D) Intracellular glutamine levels of _Tsc2_−/− MEFs: (C) stably expressing S6KCA (F5A/R5A/T389E; mutating either the three arginines or all the residues within the RSPRR motif to alanines shows same effect) [10] or empty vector, and treated with rapamycin 20ng/mL or DMSO for 48h; (D) transfected with a non-targeting control siRNA (NTC) or siRNA against both S6K1/2. 24h post-transfection, cells transfected with NTC siRNA were treated with PF4708671 10μM or DMSO for 48h. (E) Glutamine consumption of _Tsc2_−/− MEFs transfected with a non-targeting control siRNA (NTC) or siRNA against both S6K1/2. 72h post-transfection, media was collected and levels of glutamine in the media were determined. (F) Normalized luciferase light units of Tsc2 WT MEFs transfected with the pDLN reporter construct containing the 5’UTR of Myc under the control of renilla luciferase. Firefly luciferase was used as an internal control. 48h post-transfection, cells were treated with rapamycin 20ng/mL or PF4708671 10μM for 8h. (G) Relative levels of Myc, Gls and Actin mRNA in each polysomal gradient fraction. mRNA levels were measured by qPCR and normalized to 5S rRNA level. HEK293T cells were treated with rapamycin 20ng/mL for 24h, and polysomes were fractionated on sucrose density gradients. (H-I) GLS and Myc protein levels in whole cell lysates from: (H) _Tsc2_−/− MEFs transfected with a non-targeting control siRNA (NTC) or two independent siRNAs against eIF4B for 72h; (I) _Tsc2_−/− MEFs stably expressing eIF4B WT, mutant S422D or empty vector (EV) and treated with rapamycin for 24h. The mean is shown; error bars represent SEM from at least three biological replicates. The asterisk (*) denotes a nonspecific band. Numbers below the immunoblot image represent quantification normalized to the loading control. See also Figure S2 and S3.
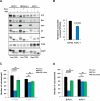
Figure 4. Inhibition of GLS reduces the growth of pancreatic cancer cells
(A) GLS and Myc protein levels in whole cell lysates from BxPC3, MIAPaCa-2 or AsPC-1 cells treated with rapamycin 20ng/mL or BEZ235 1μM for 24h. (B) Glutamine consumption of BxPC3 or AsPC-1 cells 48h after plating. (C-D) Soft agar assays with: (C) BxPC3 or AsPC-1 cells treated with BPTES (10μM), and the combination of BPTES (10μM)+OAA (2mM); (D) BxPC3 or AsPC-1 cells treated with BPTES, and the combination of BPTES (10μM)+NAC (10mM). The mean is shown; error bars represent SEM from at least three biological replicates.
References
- Howell JJ, Ricoult SJ, Ben-Sahra I, Manning BD. A growing role for mTOR in promoting anabolic metabolism. Biochem Soc Trans. 2013;41:906–912. - PubMed
- DeBerardinis RJ, Lum JJ, Hatzivassiliou G, Thompson CB. The biology of cancer: metabolic reprogramming fuels cell growth and proliferation. Cell Metabolism. 2008;7:11–20. - PubMed
- Aledo JC, Gómez-Fabre PM, Olalla L, Márquez J. Identification of two human glutaminase loci and tissue-specific expression of the two related genes. Mamm. Genome. 2010;11:1107–1110. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 HL121266/HL/NHLBI NIH HHS/United States
- R01 CA046595/CA/NCI NIH HHS/United States
- R01 GM051405/GM/NIGMS NIH HHS/United States
- GM51405/GM/NIGMS NIH HHS/United States
- R37 CA046595/CA/NCI NIH HHS/United States
- CA46595/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous