Cancer genomics: one cell at a time - PubMed (original) (raw)
Review
Cancer genomics: one cell at a time
Nicholas E Navin. Genome Biol. 2014.
Abstract
The study of single cancer cells has transformed from qualitative microscopic images to quantitative genomic datasets. This paradigm shift has been fueled by the development of single-cell sequencing technologies, which provide a powerful new approach to study complex biological processes in human cancers.
Figures
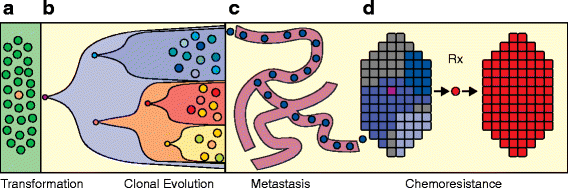
Figure 1
Single-cell processes in cancer. Although single cancer cells interact with their neighbors and the adjacent stromal cells, there are many biological processes that occur through the actions of individual cancer cells, shown in this illustration. These complex biological processes in human cancers include: (a) transformation from a single normal somatic cell into a tumor cell; (b) clonal evolution that occurs through a series of selective sweeps when single cells acquire driver mutations and diversify, leading to intratumor heterogeneity; (c) single cells from the primary tumor intravasate into the circulatory system and extravasate at distant organ sites to form metastatic tumors; and (d) the evolution of chemoresistance that occurs when the tumor is eradicated but survived by single tumor cells that harbor resistance mutations and expand to reconstitute the tumor mass.
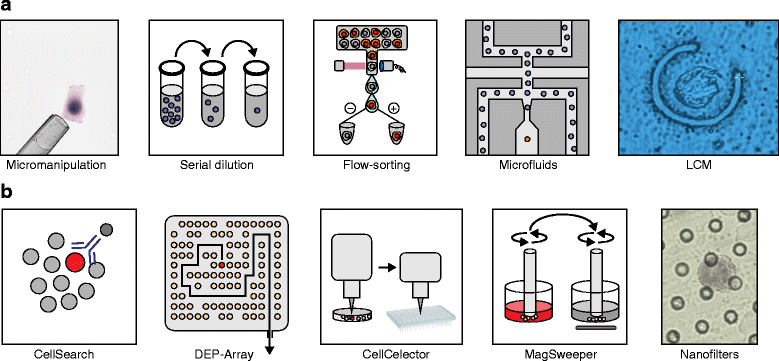
Figure 2
Methods for isolating single cancer cells from abundant and rare populations. (a) Methods for isolating single cells from abundant cellular populations include: micromanipulation by robotics or mouth pipetting, serial dilutions, flow-sorting, microfluidics platforms and laser-capture microdissection (LCM; 63X objective). (b) Methods for isolating single cells from rare cellular populations include: CellSearch (Johnson & Johnson), DEP-Array (Silicon Biosciences), CellCelector (Automated Lab Solutions), MagSweeper (Illumina) and nano-fabricated filters (Creatv MicroTech).
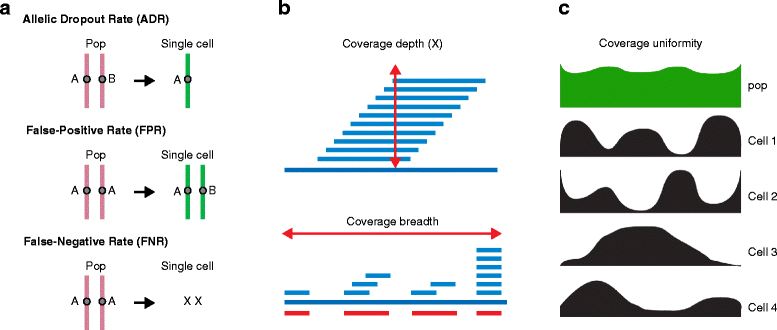
Figure 3
Technical errors and coverage in single-cell sequencing data. (a) Technical errors that occur in single-cell sequencing (SCS) data include: false-positive errors, allelic dropout events and false-negative errors due to insufficient coverage. ‘Pop’ indicates a population of cells. (b) Coverage metrics in SCS data include coverage depth and total physical coverage, or breadth. (c) Coverage uniformity, or ‘eveness’ in SCS data can vary from cell to cell, but is often more uniform in standard genomic DNA sequencing experiments using populations of cells.
Similar articles
- Measuring Clonal Evolution in Cancer with Genomics.
Williams MJ, Sottoriva A, Graham TA. Williams MJ, et al. Annu Rev Genomics Hum Genet. 2019 Aug 31;20:309-329. doi: 10.1146/annurev-genom-083117-021712. Epub 2019 May 5. Annu Rev Genomics Hum Genet. 2019. PMID: 31059289 Review. - Advances for studying clonal evolution in cancer.
Ding L, Raphael BJ, Chen F, Wendl MC. Ding L, et al. Cancer Lett. 2013 Nov 1;340(2):212-9. doi: 10.1016/j.canlet.2012.12.028. Epub 2013 Jan 23. Cancer Lett. 2013. PMID: 23353056 Free PMC article. Review. - Enhancing cancer clonality analysis with integrative genomics.
Peterson EA, Bauer MA, Chavan SS, Ashby C, Weinhold N, Heuck CJ, Morgan GJ, Johann DJ. Peterson EA, et al. BMC Bioinformatics. 2015;16 Suppl 13(Suppl 13):S7. doi: 10.1186/1471-2105-16-S13-S7. Epub 2015 Sep 25. BMC Bioinformatics. 2015. PMID: 26424171 Free PMC article. - Cancer genomics: integrating form and function.
Kim SY, Hahn WC. Kim SY, et al. Carcinogenesis. 2007 Jul;28(7):1387-92. doi: 10.1093/carcin/bgm086. Epub 2007 Apr 13. Carcinogenesis. 2007. PMID: 17434922 Review. - Predictive genomics: a cancer hallmark network framework for predicting tumor clinical phenotypes using genome sequencing data.
Wang E, Zaman N, Mcgee S, Milanese JS, Masoudi-Nejad A, O'Connor-McCourt M. Wang E, et al. Semin Cancer Biol. 2015 Feb;30:4-12. doi: 10.1016/j.semcancer.2014.04.002. Epub 2014 Apr 18. Semin Cancer Biol. 2015. PMID: 24747696 Review.
Cited by
- Moss enables high sensitivity single-nucleotide variant calling from multiple bulk DNA tumor samples.
Zhang C, El-Kebir M, Ochoa I. Zhang C, et al. Nat Commun. 2021 Apr 13;12(1):2204. doi: 10.1038/s41467-021-22466-9. Nat Commun. 2021. PMID: 33850139 Free PMC article. - Genetic heterogeneity in cholangiocarcinoma: a major challenge for targeted therapies.
Brandi G, Farioli A, Astolfi A, Biasco G, Tavolari S. Brandi G, et al. Oncotarget. 2015 Jun 20;6(17):14744-53. doi: 10.18632/oncotarget.4539. Oncotarget. 2015. PMID: 26142706 Free PMC article. Review. - Exploring Current Challenges and Perspectives for Automatic Reconstruction of Clonal Evolution.
Sandmann S, Richter S, Jiang X, Varghese J. Sandmann S, et al. Cancer Genomics Proteomics. 2022 Mar-Apr;19(2):194-204. doi: 10.21873/cgp.20314. Cancer Genomics Proteomics. 2022. PMID: 35181588 Free PMC article. - Understanding tumor ecosystems by single-cell sequencing: promises and limitations.
Ren X, Kang B, Zhang Z. Ren X, et al. Genome Biol. 2018 Dec 3;19(1):211. doi: 10.1186/s13059-018-1593-z. Genome Biol. 2018. PMID: 30509292 Free PMC article. Review. - CNAsim: improved simulation of single-cell copy number profiles and DNA-seq data from tumors.
Weiner S, Bansal MS. Weiner S, et al. Bioinformatics. 2023 Jul 1;39(7):btad434. doi: 10.1093/bioinformatics/btad434. Bioinformatics. 2023. PMID: 37449891 Free PMC article.
References
- Fiegl M, Tueni C, Schenk T, Jakesz R, Gnant M, Reiner A, Rudas M, Pirc-Danoewinata H, Marosi C, Huber H, Drach J. Interphase cytogenetics reveals a high incidence of aneuploidy and intra-tumour heterogeneity in breast cancer. Br J Cancer. 1995;72:51–55. doi: 10.1038/bjc.1995.276. - DOI - PMC - PubMed
- Nik-Zainal S, Van Loo P, Wedge DC, Alexandrov LB, Greenman CD, Lau KW, Raine K, Jones D, Marshall J, Ramakrishna M, Shlien A, Cooke SL, Hinton J, Menzies A, Stebbings LA, Leroy C, Jia M, Rance R, Mudie LJ, Gamble SJ, Stephens PJ, McLaren S, Tarpey PS, Papaemmanuil E, Davies HR, Varela I, McBride DJ, Bignell GR, Leung K, Butler AP, et al. The life history of 21 breast cancers. Cell. 2012;149:994–1007. doi: 10.1016/j.cell.2012.04.023. - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
- R01 CA169244/CA/NCI NIH HHS/United States
- R21 CA174397/CA/NCI NIH HHS/United States
- R21CA174397-01/CA/NCI NIH HHS/United States
- 1R01CA169244-01/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources