The contribution of genetic variants to disease depends on the ruler - PubMed (original) (raw)
Review
. 2014 Nov;15(11):765-76.
doi: 10.1038/nrg3786. Epub 2014 Sep 16.
Affiliations
- PMID: 25223781
- PMCID: PMC4412738
- DOI: 10.1038/nrg3786
Review
The contribution of genetic variants to disease depends on the ruler
John S Witte et al. Nat Rev Genet. 2014 Nov.
Abstract
Our understanding of the genetic basis of disease has evolved from descriptions of overall heritability or familiality to the identification of large numbers of risk loci. One can quantify the impact of such loci on disease using a plethora of measures, which can guide future research decisions. However, different measures can attribute varying degrees of importance to a variant. In this Analysis, we consider and contrast the most commonly used measures - specifically, the heritability of disease liability, approximate heritability, sibling recurrence risk, overall genetic variance using a logarithmic relative risk scale, the area under the receiver-operating curve for risk prediction and the population attributable fraction - and give guidelines for their use that should be explicitly considered when assessing the contribution of genetic variants to disease.
Conflict of interest statement
Competing interests
The authors declare no competing interests.
Figures
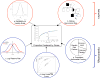
Figure 1. Different measures of genetic effects on disease
A number of different measures can be used to assess how much known genetic factors contribute to the overall genetic variation in disease. These include: a. heritability, b. sibling relative risk, c. log relative risk genetic variance, d. area under the receiver operating curve (AUC), and e. population attributable fraction. These measures have their bases in traditionally distinct disciplines such as quantitative genetics and epidemiology, which have recently begun to coalesce. While the latter were originally developed to address different questions, they are presently being repurposed to assess how much genetic variation cab be explained. We compare these measures via simulation and applications.
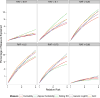
Figure 2. Empirical evaluation of measures of genetic effects
Comparison of heritability, approximate heritability, sibling relative risk, log relative risk genetic variance, and area under the curve (AUC) explained across a range of complex disease architectures. The measures are calculated for a single causal variant with risk allele frequency (RAF) = 0.01, 0.10, 0.25, 0.50, 0.75, and 0.99 and genetic relative risk (RR) ranging from 1.0 to 3.0 (assuming multiplicative model). The overall disease risk is assumed = 0.01, and the total sibling relative risk = 5, which gives an overall genetic heritability on the liability scale = 0.55 and a maximum AUC = 0.95. The percentage of heritability, sibling risk, and logRR genetic variance explained is quite modest for low RRs and small RAF, but as these increase the measures start to materially differ. Heritability is always one of the smallest measures, and is overestimated by the approximate heritability as the RR increases. The sibling relative risk and AUC are generally the largest measures for lower RAFs.
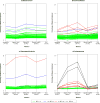
Figure 3. Application of measures to four diseases
Comparison of commonly used measures for assessing the impact of known risk variants on four diseases: a. breast cancer (65 variants), b. Crohn’s disease (143 variants), c. rheumatoid arthritis (36 variants), and d. schizophrenia (32 variants). The measures are: heritability explained; approximation of heritability explained; sibling recurrence risk explained; logRR genetic variance explained; and the proportion of area under the curve (pAUC). Each line corresponds to an individual risk variant, indicating the percentage of each measure (e.g., total variability) it explains. Lines are different colors depending on the relative risk (estimated by the odds ratio, OR) for each variant. The percentage axes are on a squared scale.
Figure 4. Aspects of disease heritability: known, hiding, and missing
A growing proportion of the total heritability estimated from family studies can be explained by known variants detected in existing genome-wide association studies (bottom). This is one of the key measures considered here. The remaining heritability can be broken into that which is ‘hiding’ versus ‘still missing’. The hiding heritability can be estimated from genome-wide arrays using the Genetic Relatedness Estimation through Maximum Likelihood (GREML) model. The still-missing heritability is that which may remain even after genome-wide association studies, reflecting for example genetic different architectures (e.g., rare variants). Note that the total heritability may be biased upward due to confounding by non-additive genetic or non-genetic factors.
Similar articles
- Measuring missing heritability: inferring the contribution of common variants.
Golan D, Lander ES, Rosset S. Golan D, et al. Proc Natl Acad Sci U S A. 2014 Dec 9;111(49):E5272-81. doi: 10.1073/pnas.1419064111. Epub 2014 Nov 24. Proc Natl Acad Sci U S A. 2014. PMID: 25422463 Free PMC article. - On the relationship between the heritability and the attributable fraction.
Dahlqwist E, Magnusson PKE, Pawitan Y, Sjölander A. Dahlqwist E, et al. Hum Genet. 2019 Apr;138(4):425-435. doi: 10.1007/s00439-019-02006-8. Epub 2019 Apr 2. Hum Genet. 2019. PMID: 30941497 Free PMC article. - Contribution of Common Genetic Variants to Familial Aggregation of Disease and Implications for Sequencing Studies.
Schlafly A, Pfeiffer RM, Nagore E, Puig S, Calista D, Ghiorzo P, Menin C, Fargnoli MC, Peris K, Song L, Zhang T, Shi J, Landi MT, Sampson JN. Schlafly A, et al. PLoS Genet. 2019 Nov 15;15(11):e1008490. doi: 10.1371/journal.pgen.1008490. eCollection 2019 Nov. PLoS Genet. 2019. PMID: 31730655 Free PMC article. - Genome-wide association studies for complex traits: consensus, uncertainty and challenges.
McCarthy MI, Abecasis GR, Cardon LR, Goldstein DB, Little J, Ioannidis JP, Hirschhorn JN. McCarthy MI, et al. Nat Rev Genet. 2008 May;9(5):356-69. doi: 10.1038/nrg2344. Nat Rev Genet. 2008. PMID: 18398418 Review. - Genetic architecture: the shape of the genetic contribution to human traits and disease.
Timpson NJ, Greenwood CMT, Soranzo N, Lawson DJ, Richards JB. Timpson NJ, et al. Nat Rev Genet. 2018 Feb;19(2):110-124. doi: 10.1038/nrg.2017.101. Epub 2017 Dec 11. Nat Rev Genet. 2018. PMID: 29225335 Review.
Cited by
- Variants associating with uterine leiomyoma highlight genetic background shared by various cancers and hormone-related traits.
Rafnar T, Gunnarsson B, Stefansson OA, Sulem P, Ingason A, Frigge ML, Stefansdottir L, Sigurdsson JK, Tragante V, Steinthorsdottir V, Styrkarsdottir U, Stacey SN, Gudmundsson J, Arnadottir GA, Oddsson A, Zink F, Halldorsson G, Sveinbjornsson G, Kristjansson RP, Davidsson OB, Salvarsdottir A, Thoroddsen A, Helgadottir EA, Kristjansdottir K, Ingthorsson O, Gudmundsson V, Geirsson RT, Arnadottir R, Gudbjartsson DF, Masson G, Asselbergs FW, Jonasson JG, Olafsson K, Thorsteinsdottir U, Halldorsson BV, Thorleifsson G, Stefansson K. Rafnar T, et al. Nat Commun. 2018 Sep 7;9(1):3636. doi: 10.1038/s41467-018-05428-6. Nat Commun. 2018. PMID: 30194396 Free PMC article. - Genome-wide meta-analysis and replication studies in multiple ethnicities identify novel adolescent idiopathic scoliosis susceptibility loci.
Khanshour AM, Kou I, Fan Y, Einarsdottir E, Makki N, Kidane YH, Kere J, Grauers A, Johnson TA, Paria N, Patel C, Singhania R, Kamiya N, Takeda K, Otomo N, Watanabe K, Luk KDK, Cheung KMC, Herring JA, Rios JJ, Ahituv N, Gerdhem P, Gurnett CA, Song YQ, Ikegawa S, Wise CA. Khanshour AM, et al. Hum Mol Genet. 2018 Nov 15;27(22):3986-3998. doi: 10.1093/hmg/ddy306. Hum Mol Genet. 2018. PMID: 30395268 Free PMC article. - Estimation of metabolic syndrome heritability in three large populations including full pedigree and genomic information.
Graziano F, Biino G, Bonati MT, Neale BM, Do R, Concas MP, Vaccargiu S, Pirastu M, Terradura-Vagnarelli O, Cirillo M, Laurenzi M, Mancini M, Zanchetti A, Grassi M. Graziano F, et al. Hum Genet. 2019 Jul;138(7):739-748. doi: 10.1007/s00439-019-02024-6. Epub 2019 Jun 1. Hum Genet. 2019. PMID: 31154530 - Cumulative Genetic Score and C9orf72 Repeat Status Independently Contribute to Amyotrophic Lateral Sclerosis Risk in 2 Case-Control Studies.
Dou J, Bakulski K, Guo K, Hur J, Zhao L, Saez-Atienzar S, Stark A, Chia R, García-Redondo A, Rojas-Garcia R, Vázquez Costa JF, Fernandez Santiago R, Bandres-Ciga S, Gómez-Garre P, Periñán MT, Mir P, Pérez-Tur J, Cardona F, Menendez-Gonzalez M, Riancho J, Borrego-Hernández D, Galán-Dávila L, Infante Ceberio J, Pastor P, Paradas C, Dols-Icardo O, Traynor BJ, Feldman EL, Goutman SA; Spanish Neurological Consortium. Dou J, et al. Neurol Genet. 2023 May 31;9(4):e200079. doi: 10.1212/NXG.0000000000200079. eCollection 2023 Aug. Neurol Genet. 2023. PMID: 37293291 Free PMC article. - HiSeeker: Detecting High-Order SNP Interactions Based on Pairwise SNP Combinations.
Liu J, Yu G, Jiang Y, Wang J. Liu J, et al. Genes (Basel). 2017 May 31;8(6):153. doi: 10.3390/genes8060153. Genes (Basel). 2017. PMID: 28561745 Free PMC article.
References
Explores the relationship between heritability on disease and liability scales
- Falconer D. The inheritance of liability to certain diseases, estimates from the incidence among relatives. Annals of Human Genetics. 1965;29:51–76.
A formal derivation of the relationship between disease risk in relatives and heritability plus a thoughtful exploration of scenarios and caveats
- Falconer D, Mackay TF. Introduction to Quantitative Genetics. Pearson Education Ltd; Harlow, England: 1996.
- Risch NJ. Searching for genetic determinants in the new millennium. Nature. 2000;405:847–56. - PubMed
Explains variance explained by a single locus on the disease and liability scale
- James JW. Frequency in relatives for an all-or-none trait. Ann Hum Genet. 1971;35:47–9. - PubMed
- Pharoah PD, et al. Polygenic susceptibility to breast cancer and implications for prevention. Nat Genet. 2002;31:33–6. - PubMed
A clear presentation of the log relative risk model
- Pharoah PD, Antoniou AC, Easton DF, Ponder BA. Polygenes, risk prediction, and targeted prevention of breast cancer. N Engl J Med. 2008;358:2796–803. - PubMed
- Pharoah PD, Day NE, Duffy S, Easton DF, Ponder BA. Family history and the risk of breast cancer: a systematic review and meta-analysis. Int J Cancer. 1997;71:800–9. - PubMed
Considers the limitations of the population attributable fraction
- Wray NR, et al. Polygenic methods and their application to psychiatric traits. Journal of Childhood Psychology and Psychiatry. 2014 - PubMed
Utilises variance explained by loci and also considers complications of age-related risk
Publication types
MeSH terms
Grants and funding
- P30 CA82103/CA/NCI NIH HHS/United States
- U01 CA127298/CA/NCI NIH HHS/United States
- R25 CA112355/CA/NCI NIH HHS/United States
- P30 CA082103/CA/NCI NIH HHS/United States
- U01 GM061390/GM/NIGMS NIH HHS/United States
- U19 GM061390/GM/NIGMS NIH HHS/United States
- R01 CA088164/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical