Probing the disordered domain of the nuclear pore complex through coarse-grained molecular dynamics simulations - PubMed (original) (raw)
Probing the disordered domain of the nuclear pore complex through coarse-grained molecular dynamics simulations
Ali Ghavami et al. Biophys J. 2014.
Abstract
The distribution of disordered proteins (FG-nups) that line the transport channel of the nuclear pore complex (NPC) is investigated by means of coarse-grained molecular dynamics simulations. A one-bead-per-amino-acid model is presented that accounts for the hydrophobic/hydrophilic and electrostatic interactions between different amino acids, polarity of the solvent, and screening of free ions. The results indicate that the interaction of the FG-nups forms a high-density, doughnut-like distribution inside the NPC, which is rich in FG-repeats. We show that the obtained distribution is encoded in the amino-acid sequence of the FG-nups and is driven by both electrostatic and hydrophobic interactions. To explore the relation between structure and function, we have systematically removed different combinations of FG-nups from the pore to simulate inviable and viable NPCs that were previously studied experimentally. The obtained density distributions show that the maximum density of the FG-nups inside the pore does not exceed 185 mg/mL in the inviable NPCs, whereas for the wild-type and viable NPCs, this value increases to 300 mg/mL. Interestingly, this maximum density is not correlated to the total mass of the FG-nups, but depends sensitively on the specific combination of essential Nups located in the central plane of the NPC.
Copyright © 2014 Biophysical Society. Published by Elsevier Inc. All rights reserved.
Figures
Figure 1
The predicted and experimental R s values for FG-nup segments plotted against the charge to hydrophobicity ratio (C/H). To see this figure in color, go online.
Figure 2
(A) Simplified geometry of the core scaffold of the yeast nuclear pore complex reconstructed based on the model of Alber et al. (42,43). The outer radius of the scaffold changes from 30 nm at the center to 33.5 nm at the peripheries. The inner blobs, which are decorated in eightfold rotational symmetry, represent Nup188 of the inner rings. The blobs at the cytoplasmic ring represent Nup82 and Nic96, whereas the ones at the nuclear ring represent Nic96. (B) The circumferential projection of radial and axial positions of the anchor points of the FG-nups. (C) The distribution of charged amino acids (red sticks) and FG-repeats (green sticks) in the sequence of the FG-regions of the FG-nups as used in the model. All Nups are anchored to the scaffold at their C-terminus. To see this figure in color, go online.
Figure 3
The three-dimensional density distribution of different amino acids inside the NPC obtained from the wild-type-1 simulation. (A) The distribution of all amino acids. The iso-surface plot corresponds to a mass density of 140 mg/mL. (B) The density distribution of charged amino acids. The iso-surface plot corresponds to a mass density of 22 mg/mL. (C) The density distribution of FG-repeats inside the transport channel of the NPC. The iso-surface plot corresponds to an average distance of 2.7 nm between the FG-repeats. To see this figure in color, go online.
Figure 4
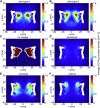
Two-dimensional density plots of the FG-nups in the simulated NPCs. (A) Wild-type-1 NPC. (B) Wild-type-2 NPC, simulated with a different starting configuration and initial velocity distribution compared to wild-type-1. (C) No-charge NPC, in which all charged residues are replaced by neutral beads in the sequence of the FG-nups. (D) Denatured NPC, where all residues are replaced by neutral beads. (E) Reversed NPC, where the FG-nups are anchored from their N-terminus. (F) Uniform NPC, in which the sequence of the FG-nups is modified such that they have a uniform distribution of charged and hydrophobic amino acids along their length. To see this figure in color, go online.
Figure 5
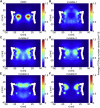
The two-dimensional density plots for viable (A) and inviable NPCs (B–F) corresponding to Table 1. To see this figure in color, go online.
Figure 6
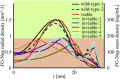
Comparison of the radial density distribution at the z location of maximal density in the two-dimensional density plots for the simulated NPCs. The mass density is calculated using an average mass of 120 Da per residue. To see this figure in color, go online.
Similar articles
- The Effect of FG-Nup Phosphorylation on NPC Selectivity: A One-Bead-Per-Amino-Acid Molecular Dynamics Study.
Mishra A, Sipma W, Veenhoff LM, Van der Giessen E, Onck PR. Mishra A, et al. Int J Mol Sci. 2019 Jan 30;20(3):596. doi: 10.3390/ijms20030596. Int J Mol Sci. 2019. PMID: 30704069 Free PMC article. - Nucleoporin's Like Charge Regions Are Major Regulators of FG Coverage and Dynamics Inside the Nuclear Pore Complex.
Peyro M, Soheilypour M, Ghavami A, Mofrad MR. Peyro M, et al. PLoS One. 2015 Dec 11;10(12):e0143745. doi: 10.1371/journal.pone.0143745. eCollection 2015. PLoS One. 2015. PMID: 26658558 Free PMC article. - Nucleoporins' exclusive amino acid sequence features regulate their transient interaction with and selectivity of cargo complexes in the nuclear pore.
Peyro M, Dickson AM, Mofrad MRK. Peyro M, et al. Mol Biol Cell. 2021 Nov 1;32(21):ar31. doi: 10.1091/mbc.E21-04-0161. Epub 2021 Sep 2. Mol Biol Cell. 2021. PMID: 34473567 Free PMC article. - Flexible gates: dynamic topologies and functions for FG nucleoporins in nucleocytoplasmic transport.
Terry LJ, Wente SR. Terry LJ, et al. Eukaryot Cell. 2009 Dec;8(12):1814-27. doi: 10.1128/EC.00225-09. Epub 2009 Oct 2. Eukaryot Cell. 2009. PMID: 19801417 Free PMC article. Review. - Phase separation of FG-nucleoporins in nuclear pore complexes.
Nag N, Sasidharan S, Uversky VN, Saudagar P, Tripathi T. Nag N, et al. Biochim Biophys Acta Mol Cell Res. 2022 Apr;1869(4):119205. doi: 10.1016/j.bbamcr.2021.119205. Epub 2022 Jan 4. Biochim Biophys Acta Mol Cell Res. 2022. PMID: 34995711 Review.
Cited by
- Spatiotemporal dynamics of the nuclear pore complex transport barrier resolved by high-speed atomic force microscopy.
Sakiyama Y, Mazur A, Kapinos LE, Lim RY. Sakiyama Y, et al. Nat Nanotechnol. 2016 Aug;11(8):719-23. doi: 10.1038/nnano.2016.62. Epub 2016 May 2. Nat Nanotechnol. 2016. PMID: 27136131 - A physical model describing the interaction of nuclear transport receptors with FG nucleoporin domain assemblies.
Zahn R, Osmanović D, Ehret S, Araya Callis C, Frey S, Stewart M, You C, Görlich D, Hoogenboom BW, Richter RP. Zahn R, et al. Elife. 2016 Apr 8;5:e14119. doi: 10.7554/eLife.14119. Elife. 2016. PMID: 27058170 Free PMC article. - Regulation of RNA-binding proteins affinity to export receptors enables the nuclear basket proteins to distinguish and retain aberrant mRNAs.
Soheilypour M, Mofrad MR. Soheilypour M, et al. Sci Rep. 2016 Nov 2;6:35380. doi: 10.1038/srep35380. Sci Rep. 2016. PMID: 27805000 Free PMC article. - The Effect of FG-Nup Phosphorylation on NPC Selectivity: A One-Bead-Per-Amino-Acid Molecular Dynamics Study.
Mishra A, Sipma W, Veenhoff LM, Van der Giessen E, Onck PR. Mishra A, et al. Int J Mol Sci. 2019 Jan 30;20(3):596. doi: 10.3390/ijms20030596. Int J Mol Sci. 2019. PMID: 30704069 Free PMC article. - A One-Bead-Per-Saccharide (1BPS) Model for Glycosaminoglycans.
Shakibi S, Onck PR, Van der Giessen E. Shakibi S, et al. J Chem Theory Comput. 2023 Aug 22;19(16):5491-5502. doi: 10.1021/acs.jctc.3c00238. Epub 2023 Jul 17. J Chem Theory Comput. 2023. PMID: 37459601 Free PMC article.
References
- Feldherr C.M., Akin D. The location of the transport gate in the nuclear pore complex. J. Cell Sci. 1997;110:3065–3070. - PubMed
- Peters R. Translocation through the nuclear pore: Kaps pave the way. BioEssays. 2009;31:466–477. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources