Genomic analysis of bone marrow failure and myelodysplastic syndromes reveals phenotypic and diagnostic complexity - PubMed (original) (raw)
Comparative Study
doi: 10.3324/haematol.2014.113456. Epub 2014 Sep 19.
Siobán B Keel 2, Tom Walsh 3, Ming K Lee 3, Suleyman Gulsuner 3, Amanda C Watts 3, Colin C Pritchard 4, Stephen J Salipante 4, Michael R Jeng 5, Inga Hofmann 6, David A Williams 7, Mark D Fleming 8, Janis L Abkowitz 2, Mary-Claire King 3, Akiko Shimamura 9
Affiliations
- PMID: 25239263
- PMCID: PMC4281311
- DOI: 10.3324/haematol.2014.113456
Comparative Study
Genomic analysis of bone marrow failure and myelodysplastic syndromes reveals phenotypic and diagnostic complexity
Michael Y Zhang et al. Haematologica. 2015 Jan.
Abstract
Accurate and timely diagnosis of inherited bone marrow failure and inherited myelodysplastic syndromes is essential to guide clinical management. Distinguishing inherited from acquired bone marrow failure/myelodysplastic syndrome poses a significant clinical challenge. At present, diagnostic genetic testing for inherited bone marrow failure/myelodysplastic syndrome is performed gene-by-gene, guided by clinical and laboratory evaluation. We hypothesized that standard clinically-directed genetic testing misses patients with cryptic or atypical presentations of inherited bone marrow failure/myelodysplastic syndrome. In order to screen simultaneously for mutations of all classes in bone marrow failure/myelodysplastic syndrome genes, we developed and validated a panel of 85 genes for targeted capture and multiplexed massively parallel sequencing. In patients with clinical diagnoses of Fanconi anemia, genomic analysis resolved subtype assignment, including those of patients with inconclusive complementation test results. Eight out of 71 patients with idiopathic bone marrow failure or myelodysplastic syndrome were found to harbor damaging germline mutations in GATA2, RUNX1, DKC1, or LIG4. All 8 of these patients lacked classical clinical stigmata or laboratory findings of these syndromes and only 4 had a family history suggestive of inherited disease. These results reflect the extensive genetic heterogeneity and phenotypic complexity of bone marrow failure/myelodysplastic syndrome phenotypes. This study supports the integration of broad unbiased genetic screening into the diagnostic workup of children and young adults with bone marrow failure and myelodysplastic syndromes.
Copyright© Ferrata Storti Foundation.
Figures
Figure 1.
Detection of genomic copy number variants. Ratios of sample to median corrected depth of coverage within a flow cell lane are plotted across targeted genomic regions of the indicated gene. Diploid bases are shown in black. Deletions and duplications are shown in red and blue, respectively. Genomic positions of exons (vertical bars) and untranslated regions (light blue rectangles) are shown above ratio plots. (A) Whole gene deletion of RUNX1. No diploid bases were present in this region. (B) Deletion of FANCA exons 9–22. (C) Amplification of FANCA exons 15–22. (D) Deletion of FANCA exon 29.
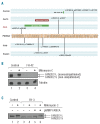
Figure 2.
Targeted gene capture correction of Fanconi anemia subtype assignment. (A) Biallelic FANCA mutations identified by MarrowSeq in 5 patients. FA patient FH-3 (highlighted in red) was non-ACG subtype by clinical complementation testing. (B) Protein extracts of bone marrow fibroblasts isolated from healthy controls or Fanconi anemia patient FH-42 (FANCD2, p.[Leu683Pro];[Glu906Ilefs*4]) were immunoblotted for FANCD2 with or without 24-h mitomycin C treatment. Fibroblasts from FH-42 exhibit low FANCD2 protein expression (lanes 3 and 4) in comparison to cells from controls (lanes 1 and 2). α-tubulin was used to ascertain equivalent protein loading. (C) Functional validation of Fanconi anemia subtype A in FA patient FH-3 (FANCA p.[Cys218Arg];[Val265Leufs*8]). Protein extracts of bone marrow fibroblasts isolated from healthy control or FH-3 were immunoblotted for FANCD2 with or without 24-h mitomycin C treatment. Fibroblasts from healthy control show both non-ubiquitinated (FANCD2-S) and monoubiquitinated (FANCD2-L) FANCD2 forms (lane 1), with an increased ratio of monoubiquitinated FANCD2 relative to non-ubiquitinated FANCD2 upon mitomycin C treatment (lane 2). Fibroblasts from FH-3 show only the non-ubiquitinated FANCD2-S form with and without mitomycin C (lanes 3 and 4). FANCD2 monoubiquitination is restored upon infection with a pMMP retroviral vector encoding the wild-type FANCA cDNA (lanes 5 and 6).
Similar articles
- Genetic features of myelodysplastic syndrome and aplastic anemia in pediatric and young adult patients.
Keel SB, Scott A, Sanchez-Bonilla M, Ho PA, Gulsuner S, Pritchard CC, Abkowitz JL, King MC, Walsh T, Shimamura A. Keel SB, et al. Haematologica. 2016 Nov;101(11):1343-1350. doi: 10.3324/haematol.2016.149476. Epub 2016 Jul 14. Haematologica. 2016. PMID: 27418648 Free PMC article. - The clinical and laboratory evaluation of patients with suspected hypocellular marrow failure.
Keel S, Geddis A. Keel S, et al. Hematology Am Soc Hematol Educ Program. 2021 Dec 10;2021(1):134-142. doi: 10.1182/hematology.2021000244. Hematology Am Soc Hematol Educ Program. 2021. PMID: 34889426 Free PMC article. Review. - Loss of B cells and their precursors is the most constant feature of GATA-2 deficiency in childhood myelodysplastic syndrome.
Nováková M, Žaliová M, Suková M, Wlodarski M, Janda A, Froňková E, Campr V, Lejhancová K, Zapletal O, Pospíšilová D, Černá Z, Kuhn T, Švec P, Pelková V, Zemanová Z, Kerndrup G, van den Heuvel-Eibrink M, van der Velden V, Niemeyer C, Kalina T, Trka J, Starý J, Hrušák O, Mejstříková E. Nováková M, et al. Haematologica. 2016 Jun;101(6):707-16. doi: 10.3324/haematol.2015.137711. Epub 2016 Mar 24. Haematologica. 2016. PMID: 27013649 Free PMC article. - A landscape of germ line mutations in a cohort of inherited bone marrow failure patients.
Bluteau O, Sebert M, Leblanc T, Peffault de Latour R, Quentin S, Lainey E, Hernandez L, Dalle JH, Sicre de Fontbrune F, Lengline E, Itzykson R, Clappier E, Boissel N, Vasquez N, Da Costa M, Masliah-Planchon J, Cuccuini W, Raimbault A, De Jaegere L, Adès L, Fenaux P, Maury S, Schmitt C, Muller M, Domenech C, Blin N, Bruno B, Pellier I, Hunault M, Blanche S, Petit A, Leverger G, Michel G, Bertrand Y, Baruchel A, Socié G, Soulier J. Bluteau O, et al. Blood. 2018 Feb 15;131(7):717-732. doi: 10.1182/blood-2017-09-806489. Epub 2017 Nov 16. Blood. 2018. PMID: 29146883 - Myelodysplastic neoplasms evolving from inherited bone marrow failure syndromes / germline predisposition syndromes: Back under the microscope.
Elghetany MT, Patnaik MM, Khoury JD. Elghetany MT, et al. Leuk Res. 2024 Feb;137:107441. doi: 10.1016/j.leukres.2024.107441. Epub 2024 Jan 23. Leuk Res. 2024. PMID: 38301422 Review.
Cited by
- Integrated proteogenomic analysis for inherited bone marrow failure syndrome.
Wakamatsu M, Muramatsu H, Sato H, Ishikawa M, Konno R, Nakajima D, Hamada M, Okuno Y, Kawashima Y, Hama A, Ito M, Iwafuchi H, Takahashi Y, Ohara O. Wakamatsu M, et al. Leukemia. 2024 Jun;38(6):1256-1265. doi: 10.1038/s41375-024-02263-1. Epub 2024 May 13. Leukemia. 2024. PMID: 38740980 Free PMC article. - Genetic Characteristics of Patients with Young-Onset Myelodysplastic Neoplasms.
Kim HY, Yoo KH, Jung CW, Kim HJ, Kim SH. Kim HY, et al. J Clin Med. 2023 Dec 13;12(24):7651. doi: 10.3390/jcm12247651. J Clin Med. 2023. PMID: 38137719 Free PMC article. - GATA2 Deficiency: Predisposition to Myeloid Malignancy and Hematopoietic Cell Transplantation.
Rajput RV, Arnold DE. Rajput RV, et al. Curr Hematol Malig Rep. 2023 Aug;18(4):89-97. doi: 10.1007/s11899-023-00695-7. Epub 2023 May 29. Curr Hematol Malig Rep. 2023. PMID: 37247092 Review. - Autoimmunity and immunodeficiency associated with monoallelic LIG4 mutations via haploinsufficiency.
Jauch AJ, Bignucolo O, Seki S, Ghraichy M, Delmonte OM, von Niederhäusern V, Higgins R, Ghosh A, Nishizawa M, Tanaka M, Baldrich A, Köppen J, Hirsiger JR, Hupfer R, Ehl S, Rensing-Ehl A, Hopfer H, Prince SS, Daley SR, Marquardsen FA, Meyer BJ, Tamm M, Daikeler TD, Diesch T, Kühne T, Helbling A, Berkemeier C, Heijnen I, Navarini AA, Trück J, de Villartay JP, Oxenius A, Berger CT, Hess C, Notarangelo LD, Yamamoto H, Recher M. Jauch AJ, et al. J Allergy Clin Immunol. 2023 Aug;152(2):500-516. doi: 10.1016/j.jaci.2023.03.022. Epub 2023 Mar 31. J Allergy Clin Immunol. 2023. PMID: 37004747 Free PMC article. - The Clinical Spectrum, Diagnosis, and Management of GATA2 Deficiency.
Santiago M, Liquori A, Such E, Zúñiga Á, Cervera J. Santiago M, et al. Cancers (Basel). 2023 Mar 3;15(5):1590. doi: 10.3390/cancers15051590. Cancers (Basel). 2023. PMID: 36900380 Free PMC article. Review.
References
- Churpek JE, Lorenz R, Nedumgottil S, Onel K, Olopade OI, Sorrell A, et al. Proposal for the clinical detection and management of patients and their family members with familial myelodysplastic syndrome/acute leukemia predisposition syndromes. Leuk Lymphoma. 2013;54(1):28–35. - PubMed
- Vardiman JW, Thiele J, Arber DA, Brunning RD, Borowitz MJ, Porwit A, et al. The 2008 revision of the World Health Organization (WHO) classification of myeloid neoplasms and acute leukemia: rationale and important changes. Blood. 2009;114(5):937–51. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R24 DK099808/DK/NIDDK NIH HHS/United States
- T32AG000057/AG/NIA NIH HHS/United States
- R24 DK093425/DK/NIDDK NIH HHS/United States
- R24 DK094746/DK/NIDDK NIH HHS/United States
- R24DK099808/DK/NIDDK NIH HHS/United States
- T32 AG000057/AG/NIA NIH HHS/United States
- R24DK094746/DK/NIDDK NIH HHS/United States
- T32 GM007266/GM/NIGMS NIH HHS/United States
- R24DK093425/DK/NIDDK NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials