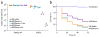Sequence-specific antimicrobials using efficiently delivered RNA-guided nucleases - PubMed (original) (raw)
. 2014 Nov;32(11):1141-5.
doi: 10.1038/nbt.3011. Epub 2014 Sep 21.
Affiliations
- PMID: 25240928
- PMCID: PMC4237163
- DOI: 10.1038/nbt.3011
Sequence-specific antimicrobials using efficiently delivered RNA-guided nucleases
Robert J Citorik et al. Nat Biotechnol. 2014 Nov.
Abstract
Current antibiotics tend to be broad spectrum, leading to indiscriminate killing of commensal bacteria and accelerated evolution of drug resistance. Here, we use CRISPR-Cas technology to create antimicrobials whose spectrum of activity is chosen by design. RNA-guided nucleases (RGNs) targeting specific DNA sequences are delivered efficiently to microbial populations using bacteriophage or bacteria carrying plasmids transmissible by conjugation. The DNA targets of RGNs can be undesirable genes or polymorphisms, including antibiotic resistance and virulence determinants in carbapenem-resistant Enterobacteriaceae and enterohemorrhagic Escherichia coli. Delivery of RGNs significantly improves survival in a Galleria mellonella infection model. We also show that RGNs enable modulation of complex bacterial populations by selective knockdown of targeted strains based on genetic signatures. RGNs constitute a class of highly discriminatory, customizable antimicrobials that enact selective pressure at the DNA level to reduce the prevalence of undesired genes, minimize off-target effects and enable programmable remodeling of microbiota.
Conflict of interest statement
COMPETING FINANCIAL INTERESTS
The authors declare competing financial interests: details are available in the online version of the paper.
Figures
Figure 1
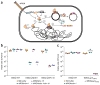
RGN constructs delivered by bacteriophage particles (ΦRGN) exhibit efficient and specific antimicrobial effects against strains harboring plasmid or chromosomal target sequences. (a) Bacteriophage-delivered RGN constructs differentially affect host cell physiology in a sequence-dependent manner. If the target sequence is: (i) absent, the RGN exerts no effect; (ii) chromosomal, RGN activity is cytotoxic; (iii) episomal, the RGN leads to either (iiia) cell death or (iiib) plasmid loss, depending on the presence or absence of toxin-antitoxin (TA) systems, respectively. (b) Treatment of EMG2 wild-type (WT) or EMG2 containing native resistance plasmids, pNDM-1 (encoding bla_NDM-1) or pSHV-18 (encoding bla_SHV-18), with SM buffer, ΦRGN_ndm-1, ΦRGN_shv-18, or multiplexed ΦRGN_ndm-1/shv-18_ at a multiplicity of infection (MOI) ~20 showed sequence-dependent cytotoxicity as evidenced by a strain-specific reduction in viable cell counts (n = 3). CFU, colony-forming units. (c) E. coli EMG2 WT or EMG2 gyrA_D87G populations were treated with SM buffer, ΦRGN_ndm-1 or ΦRGN_gyrA_D87G at MOI ~20, and viable cells were determined by plating onto Luria-Bertani agar (n = 3).
Figure 2
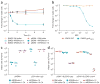
Characterization of ΦRGN-mediated killing of antibiotic-resistant bacteria. (a) Time-course treatment of EMG2 WT or EMG2 pNDM-1 with SM buffer, ΦRGN_ndm-1_ or ΦRGN_shv-18_ at a multiplicity of infection (MOI) ~20. Data represent the fold change in viable colonies at indicated time points relative to time 0 h. (b) Dose-response curve of EMG2 WT and EMG2 gyrA_D87G treated with various concentrations of ΦRGN_gyrA_D87G for 2 h. Data represent fold change in viable colonies relative to samples treated with SM buffer. Error bars (a, b), s.e.m. of three independent biological replicates (n = 3). (c) EMG2 E. coli containing the natural pNDM-1 plasmid or the bla_NDM-1 gene in a synthetic expression vector (pZA-ndm1_-gfp) were treated with either ΦRGN_ndm-1 or ΦRGN_shv-18 at MOI ~ 20 and plated onto both nonselective LB and LB + carbenicillin (Cb) to select for bla_NDM-1-containing cells. ΦRGN_ndm-1 treatment of cells harboring pNDM-1 resulted in a reduction in viability in the absence of selection, whereas ΦRGN_ndm-1 treatment of cells with pZA-ndm1_-gfp demonstrated similar cytotoxicity only under selective pressure for maintenance of the pZA-ndm1_-gfp plasmid. (d) EMG2 pSHV-18 complemented with the cognate antitoxin (pZA31-pemI) for the PemK toxin or a control vector (pZA31-gfp) was treated with SM buffer, ΦRGN_ndm-1 or ΦRGN_shv-18. Cultures were plated on LB and LB + Cb and colonies were enumerated to assess cytotoxicity or plasmid loss.
Figure 3
ΦRGN particles elicit sequence-specific toxicity against enterohemorrhagic E. coli in vitro and in vivo. (a) E. coli EMG2 wild-type (WT) cells or ATCC 43888 F′ (EHEC) cells were treated with SM buffer, ΦRGN_ndm-1_ or ΦRGN_eae_ at a multiplicity of infection (MOI) ~100 and plated onto LB agar to enumerate total cell number or LB+kanamycin (Km) to select for transductants with ΦRGNs (n = 3). (b) G. mellonella larvae were injected with either PBS or approximately 4 × 105 colony forming units (CFU) of EHEC. Subsequent administration of ΦRGN_eae_ at MOI ~30 significantly improved survival compared to SM buffer or ΦRGN_ndm-1_ treatment (Log-rank test, P < 0.001). Survival curves represent an aggregate of four independent experiments, each with 20 worms per treatment group (n = 80).
Figure 4
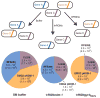
Programmable remodeling of a synthetic microbial consortium. A synthetic population composed of three different E. coli strains was treated with either SM buffer, ΦRGN_ndm-1_, or ΦRGN_gyrA_D87G at an MOI ~100 and plated onto LB with chloramphenicol, streptomycin or ofloxacin to enumerate viable cells of E. coli CJ236, EMG2 pNDM-1 or RFS289 strains, respectively. ΦRGN_ndm-1_ targets _bla_NDM-1 in EMG2 pNDM-1 and ΦRGN_gyrA_D87G targets the _gyrA_D87G allele in RFS289. Circle area is proportional to total population size and numbers represent viable cell concentrations (CFU/ml) of each strain after the indicated treatment. The s.e.m. based on three independent experiments is indicated in parentheses (n = 3).
Comment in
- A CRISPR design for next-generation antimicrobials.
Beisel CL, Gomaa AA, Barrangou R. Beisel CL, et al. Genome Biol. 2014 Nov 8;15(11):516. doi: 10.1186/s13059-014-0516-x. Genome Biol. 2014. PMID: 25417800 Free PMC article.
Similar articles
- A CRISPR design for next-generation antimicrobials.
Beisel CL, Gomaa AA, Barrangou R. Beisel CL, et al. Genome Biol. 2014 Nov 8;15(11):516. doi: 10.1186/s13059-014-0516-x. Genome Biol. 2014. PMID: 25417800 Free PMC article. - Exploiting CRISPR-Cas nucleases to produce sequence-specific antimicrobials.
Bikard D, Euler CW, Jiang W, Nussenzweig PM, Goldberg GW, Duportet X, Fischetti VA, Marraffini LA. Bikard D, et al. Nat Biotechnol. 2014 Nov;32(11):1146-50. doi: 10.1038/nbt.3043. Epub 2014 Oct 5. Nat Biotechnol. 2014. PMID: 25282355 Free PMC article. - Engineering conjugative CRISPR-Cas9 systems for the targeted control of enteric pathogens and antibiotic resistance.
Sheng H, Wu S, Xue Y, Zhao W, Caplan AB, Hovde CJ, Minnich SA. Sheng H, et al. PLoS One. 2023 Sep 12;18(9):e0291520. doi: 10.1371/journal.pone.0291520. eCollection 2023. PLoS One. 2023. PMID: 37699034 Free PMC article. - CRISPR-Cas-Based Antimicrobials: Design, Challenges, and Bacterial Mechanisms of Resistance.
Mayorga-Ramos A, Zúñiga-Miranda J, Carrera-Pacheco SE, Barba-Ostria C, Guamán LP. Mayorga-Ramos A, et al. ACS Infect Dis. 2023 Jul 14;9(7):1283-1302. doi: 10.1021/acsinfecdis.2c00649. Epub 2023 Jun 22. ACS Infect Dis. 2023. PMID: 37347230 Free PMC article. Review. - Using CRISPR-Cas systems as antimicrobials.
Bikard D, Barrangou R. Bikard D, et al. Curr Opin Microbiol. 2017 Jun;37:155-160. doi: 10.1016/j.mib.2017.08.005. Epub 2017 Sep 6. Curr Opin Microbiol. 2017. PMID: 28888103 Review.
Cited by
- The phages of staphylococci: critical catalysts in health and disease.
Hatoum-Aslan A. Hatoum-Aslan A. Trends Microbiol. 2021 Dec;29(12):1117-1129. doi: 10.1016/j.tim.2021.04.008. Epub 2021 May 21. Trends Microbiol. 2021. PMID: 34030968 Free PMC article. Review. - CRISPR-based curing and analysis of metabolic burden of cryptic plasmids in Escherichia coli Nissle 1917.
Zainuddin HS, Bai Y, Mansell TJ. Zainuddin HS, et al. Eng Life Sci. 2019 Jun 3;19(6):478-485. doi: 10.1002/elsc.201900003. eCollection 2019 Jun. Eng Life Sci. 2019. PMID: 32625025 Free PMC article. - Interference-driven spacer acquisition is dominant over naive and primed adaptation in a native CRISPR-Cas system.
Staals RH, Jackson SA, Biswas A, Brouns SJ, Brown CM, Fineran PC. Staals RH, et al. Nat Commun. 2016 Oct 3;7:12853. doi: 10.1038/ncomms12853. Nat Commun. 2016. PMID: 27694798 Free PMC article. - Cross-Regulation between Bacteria and Phages at a Posttranscriptional Level.
Altuvia S, Storz G, Papenfort K. Altuvia S, et al. Microbiol Spectr. 2018 Jul;6(4):10.1128/microbiolspec.rwr-0027-2018. doi: 10.1128/microbiolspec.RWR-0027-2018. Microbiol Spectr. 2018. PMID: 30006994 Free PMC article. Review. - Need for Laboratory Ecosystems To Unravel the Structures and Functions of Soil Microbial Communities Mediated by Chemistry.
Zhalnina K, Zengler K, Newman D, Northen TR. Zhalnina K, et al. mBio. 2018 Jul 17;9(4):e01175-18. doi: 10.1128/mBio.01175-18. mBio. 2018. PMID: 30018110 Free PMC article.
References
- Centers for Disease Control and Prevention. Antibiotic Resistance Threats in the United States. 2013
- Nordmann P, Dortet L, Poirel L. Carbapenem resistance in Enterobacteriaceae: here is the storm! Trends Mol Med. 2012;18:263–272. - PubMed
- Barrangou R, et al. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007;315:1709–1712. - PubMed
- Garneau JE, et al. The CRISPR/Cas bacterial immune system cleaves bacteriophage and plasmid DNA. Nature. 2010;468:67–71. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- 1DP2OD008435/OD/NIH HHS/United States
- HHMI/Howard Hughes Medical Institute/United States
- T32 GM008334/GM/NIGMS NIH HHS/United States
- 1P50GM098792/GM/NIGMS NIH HHS/United States
- DP2 OD008435/OD/NIH HHS/United States
- 5T32 GM008334/GM/NIGMS NIH HHS/United States
- P50 GM098792/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials