Alternating antibiotic treatments constrain evolutionary paths to multidrug resistance - PubMed (original) (raw)
Alternating antibiotic treatments constrain evolutionary paths to multidrug resistance
Seungsoo Kim et al. Proc Natl Acad Sci U S A. 2014.
Abstract
Alternating antibiotic therapy, in which pairs of drugs are cycled during treatment, has been suggested as a means to inhibit the evolution of de novo resistance while avoiding the toxicity associated with more traditional combination therapy. However, it remains unclear under which conditions and by what means such alternating treatments impede the evolution of resistance. Here, we tracked multistep evolution of resistance in replicate populations of Staphylococcus aureus during 22 d of continuously increasing single-, mixed-, and alternating-drug treatment. In all three tested drug pairs, the alternating treatment reduced the overall rate of resistance by slowing the acquisition of resistance to one of the two component drugs, sometimes as effectively as mixed treatment. This slower rate of evolution is reflected in the genome-wide mutational profiles; under alternating treatments, bacteria acquire mutations in different genes than under corresponding single-drug treatments. To test whether this observed constraint on adaptive paths reflects trade-offs in which resistance to one drug is accompanied by sensitivity to a second drug, we profiled many single-step mutants for cross-resistance. Indeed, the average cross-resistance of single-step mutants can help predict whether or not evolution was slower in alternating drugs. Together, these results show that despite the complex evolutionary landscape of multidrug resistance, alternating-drug therapy can slow evolution by constraining the mutational paths toward resistance.
Keywords: antibiotic resistance; collateral sensitivity; drug cycling; experimental evolution; multidrug therapy.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Fig. 1.
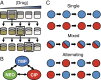
Experimental evolution of antibiotic resistance under multidrug treatments. (A) Each population of S. aureus was inoculated into a series of wells with a gradient of drug concentrations. After 20 h, the well with the highest drug concentration permitting bacterial growth (OD600 > 0.2) was used to inoculate the next cultures. This procedure was repeated for 22 d in 10 replicate populations per drug treatment. (B) Three antibiotics and their pairwise combinations were studied: TMP (blue), NEO (green), and CIP (red). (C) For each drug pair, we tested each single drug individually, a mixed treatment using a fixed ratio of the two drugs, and alternating treatments with daily switching between the two single drugs, starting with either drug.
Fig. 2.
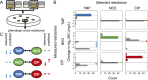
Alternating drugs slows evolution of resistance to one of the two drugs. For each of the three drug pairs, (A) TMP-NEO, (B) NEO-CIP, and (C) TMP-CIP, the mean resistance (MIC) to each of the two drugs is shown both as a function of the time exposed to that drug (Left) and as a distribution at the final time point, 11 d of exposure to that drug (Right), for the single- (TMP in blue, NEO in green, and CIP in red), alternating- (black), and mixed-drug treatments (gray; resistance to single drugs measured at only final time point). Each line indicates the mean resistance of 10 single-drug or 20 alternating-drug independent populations, as inferred from the well chosen for propagation during evolution (individual trajectories vary) (Fig. S1). Error bars indicate SEM. Day 1 represents the resistance after 2 d of evolution for half of the alternating-drug populations, and may not match that of single-drug populations. Histograms reflect phenotypic measurements following 1 d of growth in the absence of drug (Materials and Methods) (may not match exactly with inferred resistance), and triangles indicate mean final phenotyped resistance. Downward-pointing colored arrows indicate statistically significant differences (P < 0.05, two-sample t test) between the final resistance levels of the single and alternating drug treatments. In all drug pairs, resistance to one of the two drugs was slower in the alternating-drug treatment than in the single-drug treatment.
Fig. 3.
Alternating drugs constrains evolution. (A) Genes with mutations in at least four populations are shown with their frequency in each of nine treatments: trimethoprim (T, shown twice), neomycin (N), ciprofloxacin (C), their pairwise alternating treatments (double-headed arrows), and mixed treatments (plus signs). Known resistance genes are indicated at the right. (B) Genes with significant enrichment (P < 0.01) for mutations in alternating treatment compared with single drugs. Black horizontal lines indicate expected mutation frequency in alternating treatment (sum of frequencies in single drugs), and statistical significance relative to this expectation was assessed with Fisher’s exact test. *P < 0.01, **P < 0.0001. (C) Genes with significant depletion (P < 0.01) of mutations in alternating treatment. *P < 0.01. Genes with only locus tag names are indicated by the numerical portion of the locus name.
Fig. 4.
Cross-resistance explains efficacy of alternating drug treatments. (A) Single-step mutants were generated by culturing wild-type cells in liquid media with antibiotic for 20 h, then isolated by pooling wells that grew to turbidity, and then plating these mutants on agar plates with enough antibiotic to prevent growth of the wild-type cells. At least 40 resistant colonies were selected for each drug and then phenotyped in all three drugs (Materials and Methods). (B) Cross-resistance of single-step TMP-, NEO-, and CIP-resistant mutants. Histograms show the distribution of resistance level (MIC) to the three drugs relative to the ancestor. The measurements of selected resistance are shown in colors and those of cross-resistance are shown in gray. Dashed lines indicate the mean resistance. Colored arrows indicate significant cross-resistance (mean change in log2 MIC > 0.25), and colored horizontal bars indicate no significant cross-resistance (mean change in log2 MIC < 0.25). (C) Cross-resistance correctly predicts effect of alternating drugs on both component drugs for the TMP-NEO and TMP-CIP drug pairs, and on NEO for the NEO-CIP drug pair (the only inconsistency is effect on CIP in NEO-CIP drug pair). Arrows indicate change in rate of evolution of resistance in alternating drugs compared with single-drug treatment, as observed during evolution experiments (Fig. 2) and as predicted by cross-resistance measurements (B). Horizontal bars indicate a prediction or observation of no change in rate of evolution of resistance.
Comment in
- Antimicrobials: The benefits of cycling.
Nunes-Alves C. Nunes-Alves C. Nat Rev Microbiol. 2014 Nov;12(11):724-5. doi: 10.1038/nrmicro3370. Nat Rev Microbiol. 2014. PMID: 25402355 No abstract available.
Similar articles
- Evolution of the Staphylococcus argenteus ST2250 Clone in Northeastern Thailand Is Linked with the Acquisition of Livestock-Associated Staphylococcal Genes.
Moradigaravand D, Jamrozy D, Mostowy R, Anderson A, Nickerson EK, Thaipadungpanit J, Wuthiekanun V, Limmathurotsakul D, Tandhavanant S, Wikraiphat C, Wongsuvan G, Teerawattanasook N, Jutrakul Y, Srisurat N, Chaimanee P, Eoin West T, Blane B, Parkhill J, Chantratita N, Peacock SJ. Moradigaravand D, et al. mBio. 2017 Jul 5;8(4):e00802-17. doi: 10.1128/mBio.00802-17. mBio. 2017. PMID: 28679748 Free PMC article. - Collateral Resistance and Sensitivity Modulate Evolution of High-Level Resistance to Drug Combination Treatment in Staphylococcus aureus.
Rodriguez de Evgrafov M, Gumpert H, Munck C, Thomsen TT, Sommer MO. Rodriguez de Evgrafov M, et al. Mol Biol Evol. 2015 May;32(5):1175-85. doi: 10.1093/molbev/msv006. Epub 2015 Jan 23. Mol Biol Evol. 2015. PMID: 25618457 - Drug interactions modulate the potential for evolution of resistance.
Michel JB, Yeh PJ, Chait R, Moellering RC Jr, Kishony R. Michel JB, et al. Proc Natl Acad Sci U S A. 2008 Sep 30;105(39):14918-23. doi: 10.1073/pnas.0800944105. Epub 2008 Sep 24. Proc Natl Acad Sci U S A. 2008. PMID: 18815368 Free PMC article. - Collateral sensitivity of antibiotic-resistant microbes.
Pál C, Papp B, Lázár V. Pál C, et al. Trends Microbiol. 2015 Jul;23(7):401-7. doi: 10.1016/j.tim.2015.02.009. Epub 2015 Mar 25. Trends Microbiol. 2015. PMID: 25818802 Free PMC article. Review. - Interplay Between Antibiotic Resistance and Virulence During Disease Promoted by Multidrug-Resistant Bacteria.
Geisinger E, Isberg RR. Geisinger E, et al. J Infect Dis. 2017 Feb 15;215(suppl_1):S9-S17. doi: 10.1093/infdis/jiw402. J Infect Dis. 2017. PMID: 28375515 Free PMC article. Review.
Cited by
- Alternatives to antibiotics in animal agriculture: an ecoimmunological view.
Sang Y, Blecha F. Sang Y, et al. Pathogens. 2014 Dec 29;4(1):1-19. doi: 10.3390/pathogens4010001. Pathogens. 2014. PMID: 25551290 Free PMC article. Review. - Long-term effects of single and combined introductions of antibiotics and bacteriophages on populations of Pseudomonas aeruginosa.
Torres-Barceló C, Franzon B, Vasse M, Hochberg ME. Torres-Barceló C, et al. Evol Appl. 2016 Feb 18;9(4):583-95. doi: 10.1111/eva.12364. eCollection 2016 Apr. Evol Appl. 2016. PMID: 27099623 Free PMC article. - Conserved collateral antibiotic susceptibility networks in diverse clinical strains of Escherichia coli.
Podnecky NL, Fredheim EGA, Kloos J, Sørum V, Primicerio R, Roberts AP, Rozen DE, Samuelsen Ø, Johnsen PJ. Podnecky NL, et al. Nat Commun. 2018 Sep 10;9(1):3673. doi: 10.1038/s41467-018-06143-y. Nat Commun. 2018. PMID: 30202004 Free PMC article. - Burkholderia multivorans Exhibits Antibiotic Collateral Sensitivity.
Flanagan JN, Kavanaugh L, Steck TR. Flanagan JN, et al. Microb Drug Resist. 2020 Jan;26(1):1-8. doi: 10.1089/mdr.2019.0202. Epub 2019 Aug 8. Microb Drug Resist. 2020. PMID: 31393205 Free PMC article. - Delayed antibiotic exposure induces population collapse in enterococcal communities with drug-resistant subpopulations.
Hallinen KM, Karslake J, Wood KB. Hallinen KM, et al. Elife. 2020 Mar 24;9:e52813. doi: 10.7554/eLife.52813. Elife. 2020. PMID: 32207406 Free PMC article.
References
- Taubes G. The bacteria fight back. Science. 2008;321(5887):356–361. - PubMed
- Levy SB, Marshall B. Antibacterial resistance worldwide: Causes, challenges and responses. Nat Med. 2004;10(12) Suppl:S122–S129. - PubMed
- US Department of Health and Human Services . Antibiotic Resistance Threats in the United States 2013. Atlanta: Centers for Disease Control and Prevention; 2013.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical